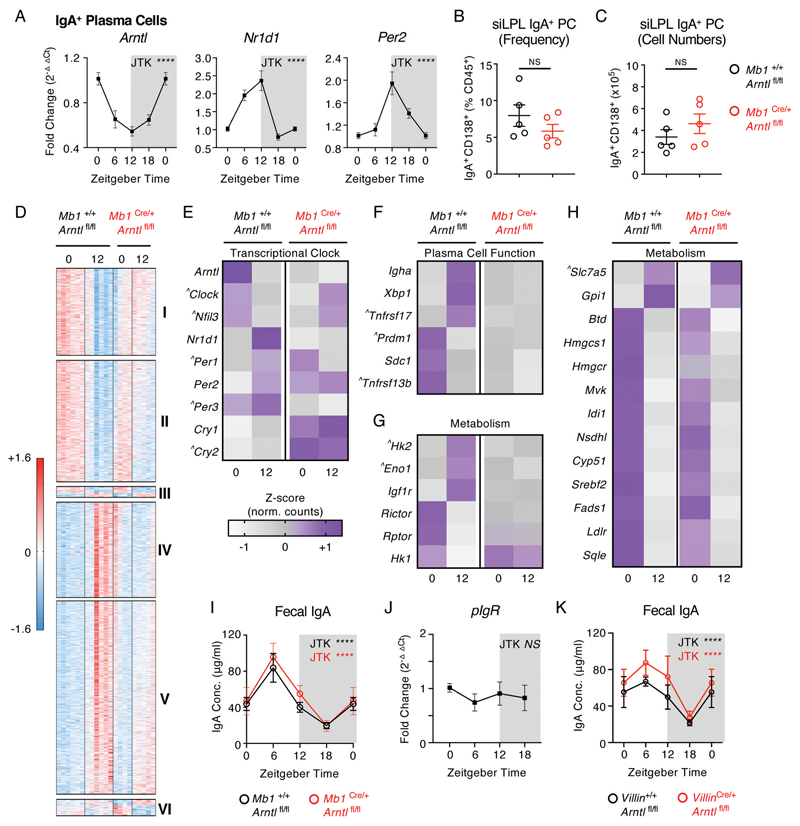
Figure 2. Rhythmic IgA+ Plasma Cell activity is in part dictated by the cell-intrinsic circadian clock.
A) Relative expression of circadian clock genes in sort-purified small intestinal IgA+ PC at ZT 0, 6, 12 and 18 (ZT0 double plotted), determined by RT-PCR. n=10 (pooled from two independent experimental cohorts). Data representative of at least 3 independent experiments. B) Frequency and C) numbers of small intestinal IgA+ PC in Mb1Cre/+ x Arntlfl/fl mice in comparison to Mb1+/+ x Arntlfl/fl littermate control animals, n=5 representative of two independent experiments. D) Heatmap comparison of significantly differentially expressed genes (Benjami-Hochberg adjusted p<0.05) identified by bulk RNA sequencing of sort-purified small intestinal IgA+ PC at ZT0 and 12, and found to significantly differ between ZT0 and ZT12 in control animals. Z-scores of relative gene expression (fpkm) values in individual animals of n=6 Mb1+/+ x Arntlfl/fl mice and n=4-5 Mb1Cre/+ x Arntlfl/fl mice per timepoint. Gene clusters: I+II (decrease in gene expression between ZT0 + ZT12 in controls, loss of suppression in Mb1Cre/+ x Arntlfl/fl mice), III (time of day difference retained in both genotypes), IV+V (increase in gene expression between ZT0 + ZT12 in controls, loss of suppression in Mb1Cre/+ x Arntlfl/fl mice) and VI (enhanced time of day difference in Mb1Cre/+ x Arntlfl/fl mice). E-H) Average z-score values (representative of n=6 Mb1+/+ x Arntlfl/fl mice and n=4-5 Mb1Cre/+ x Arntlfl/fl mice) in IgA+ PC at ZT0 and ZT12, in respect to E) Circadian clock genes, F) Plasma Cell-associated genes, G+H) Metabolism-associated genes. ^ identifies genes where time of day differences either did not reach statistical significance in control animals in this analysis but were either previously identified in Figure 1 as oscillatory, or are directly related and relevant to the biological pathway described. I) Serial fecal sampling of Mb1Cre/+ x Arntlfl/fl mice and Mb1+/+ x Arntlfl/fl mice at four time points over a circadian day (ZT 0, 6, 12, 18; ZT0 double plotted), n=8-9 and pooled from two independent experimental cohorts. J) RT-PCR expression of pIgR relative to housekeeping gene in whole small intestinal tissue samples (n=5 mice, represenative of two indepednent experiments). K) Serial fecal sampling of VillinCre/+ x Arntlfl/fl mice and Villin+/+ x Arntlfl/fl mice at four time points over a circadian day (ZT 0, 6, 12, 18; ZT0 double plotted), n=5 and representative of two independent experiments. P values for panels A and I-K were determined using JTK Cycle, D-H with Benjami-Hochberg test (DESeq2, see also methods) and panels B+C which with a parametric, unpaired t-test. All data shown as +/- SEM unless otherwise indicated, * p< 0.05, ** p< 0.01, *** p< 0.001, **** p< 0.0001.