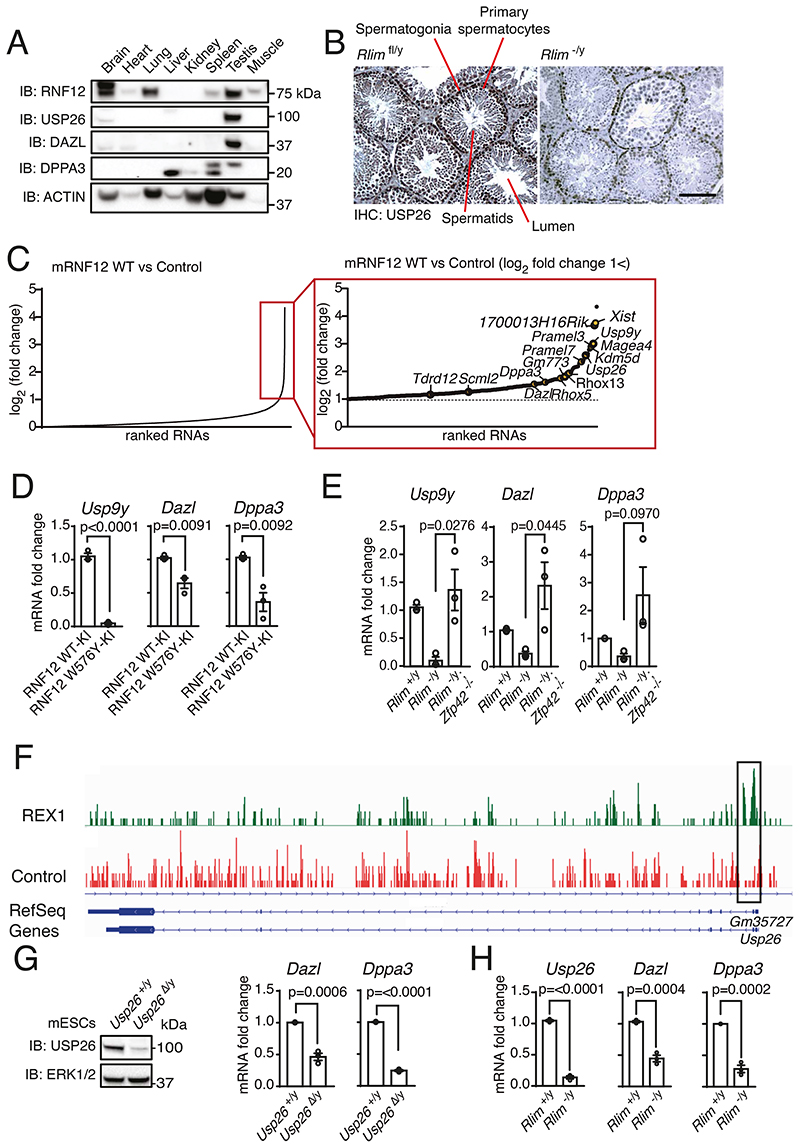
Fig. 5. The RNF12-REX1-USP26 signaling axis operates in testes and promotes expression of germ cell–specific genes.
(A) RNF12, USP26, DAZL, and DPPA3 were detected in the indicated mouse tissues by immunoblotting (IB). Actin is a loading control. Data are representative of n = 3 independent experiments. (B) USP26 immunohistochemistry of mouse testes sections from Rlimfl/y and Rlim-/y littermates. Scale bar, 100 μm. Data are representative of n = 2 animals of each genotype. (C) RNA-sequencing (RNA-SEQ) data from Rlim-/y mESCs expressing empty vector (Control) or WT-mRNF12 (17). RNAs were ranked according to fold-change increase in expression. The magnified view of the boxed area shows RNAs for which expression increased >2-fold in Rlim-/y mESCs expressing mRNF12, with RNAs implicated in reproduction or germ cell development highlighted. (D and E) Quantification of Usp9y, Dazl, and Dppa3 mRNAs in RNF12 WT-KI and W576Y-KI mESCs (D) and in Rlim-/y, Rlim-/y and Rlim✓y; Zfp42-/- mESCs (E) by qRT-PCR. Data represented as mean ± S.E.M. n = 3 independent experiments. Statistical significance was determined by paired T-test; confidence level 95%. (F) REX1 and control chromatin immunoprecipitation followed by DNA sequencing peaks for the Usp26 gene. REX1-enriched regions near the Usp26 transcriptional start site are highlighted. (G) Usp26+/y and Usp26Δ/y mESCs were cultured in 2i conditions. USP26 and ERK1/2 (loading control) were detected by immunoblotting, and Dazl and Dppa3 mRNAs were quantified by qRT-PCR. Data represented as mean ± S.E.M. n = 3 independent experiments. Statistical significance was determined by paired T-test; confidence level 95%. (H) Quantification of the indicated mRNAs in Rlim+/y and Rlim-/y in 2i mESCs by qRT-PCR. Data represented as mean ± S.E.M. n = 3 independent experiments. Statistical significance was determined by paired T-test; confidence level 95%.