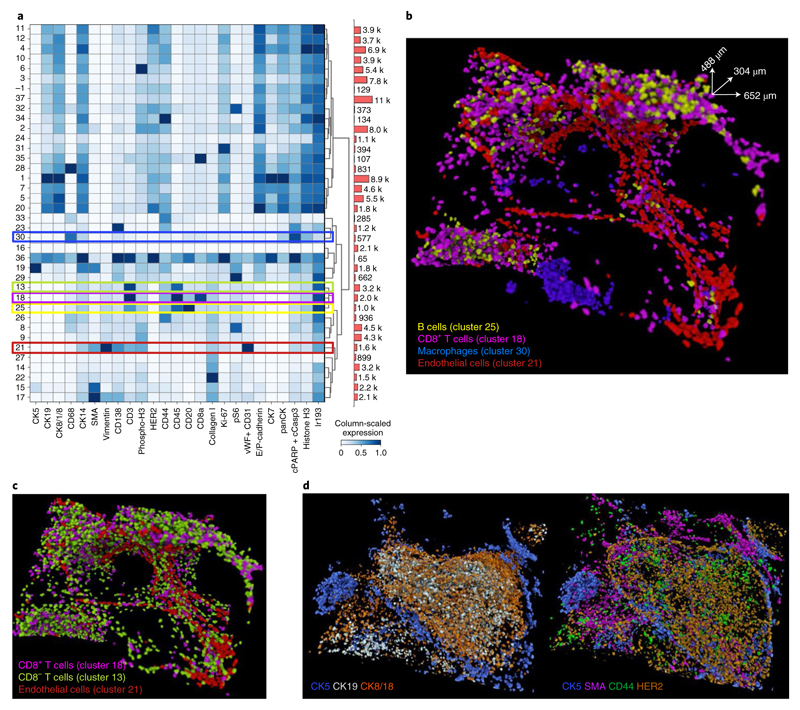
Fig. 3. Single-cell analysis of 3D IMC data in a breast carcinoma sample.
a, Heat map of an average marker expression (x axis) for each phenotypic cluster (y axis) after measuring the raw data for each segmented cell and clustering the cells using Phenograph from one out of the two breast carcinoma 3D IMC models used in this study (152 slices) (left). Marker expression data for each cell were calculated as the mean intensities of ion counts over the object mask. Colored bars indicate clusters corresponding to specific cell types, color coded as in c and d. Before clustering, data were range normalized to 99th percentile and z-scored for heat map visualization. Absolute cell counts for each cluster (right). b, 3D rendering of cells belonging to cluster 21 (endothelial cells), cluster 18 (CD8a+ T cells), cluster 25 (B cells) and cluster 30 (macrophages). c, 3D rendering of cells belonging to cluster 21 (vWF+CD31+), cluster 18 (CD8+, CD3+ and CD45+) and cluster 13 (CD8a−, CD3+ and CD45+). d, Single-cell marker expression for CK5, CK19, CK8/18, HER2, CD44 and SMA for each individual cell, where each voxel for every cell is assigned its marker expression value. All renders in this figure are representative images from one of two breast cancer 3D IMC models and are displayed in the same orientation as in Fig. 1b. AGAVE 1.0.0.1 was used for 3D rendering of the data.