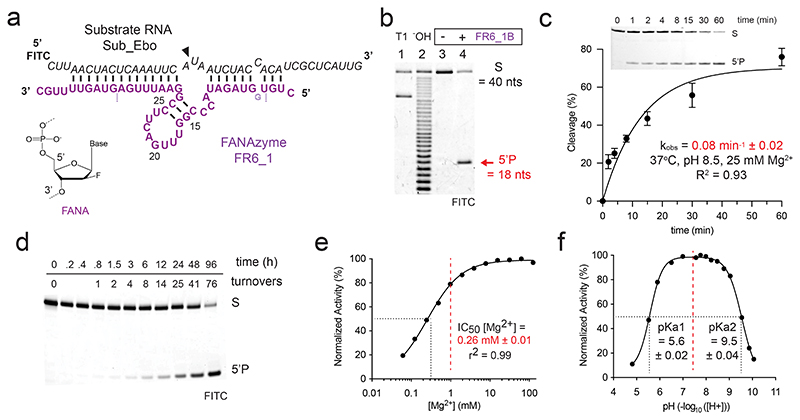
Figure 1. FR6_1 is an RNA endonuclease XNAzyme composed of 2’-deoxy-2’-fluoro-β-D-arabino nucleic acids (FANA).
(a) Sequence and putative secondary structure of XNAzyme “FR6_1” selected to target RNA “Sub_Ebo” (residues 16062-16101 of the Ebola Zaire virus genome). FANA nucleotides are shown in purple, RNA in black. Cleavage site is denoted by (▼). Dashed lines denote termini of version used for reactions in trans (“FR6_1B”), which also included an additional FANA-G between residues 4 and 5 as shown. Numbering is based on the parental FR6_1 sequence. (b) Urea-PAGE gel showing substrate RNA Sub_Ebo (1 uM) incubated with or without XNAzyme FR6_1B (5 uM) in trans (1 h, 17 °C, 10 mM Mg2+). “T1” and “-OH” indicate hydrolysis of RNA Sub_Ebo with RNase T1 (cleaves at guanine residues) or alkaline conditions, respectively, as reference. (c) Pre-steady state bimolecular reaction of substrate RNA Sub_Ebo (1 uM) with XNAzyme FR6_1B (5 uM)(37 °C, pH 8.5, 25 mM Mg2+). Error bars show standard error of the mean (s.e.m.) of three independent replicates. (d) Urea-PAGE gel showing FR6_1B (10 nM) performing multiple turnovers of Sub_Ebo (1 uM) RNA cleavage. Normalised activity of FR6_1B (5 uM) on Sub_Ebo RNA (1 uM) in varying (e) concentrations of MgCl2 or (f) pH, showing that FR6_1 retains the majority of its function at physiological Mg2+ and pH (dashed red lines).