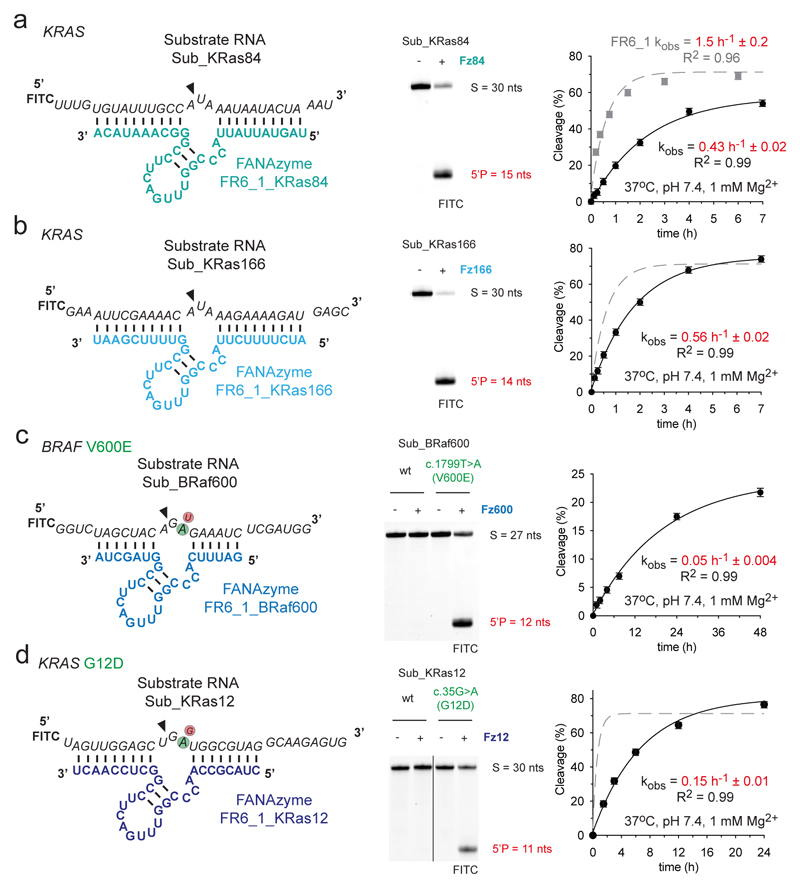
Figure 2. FR6_1 is a modular XNAzyme and can be re-programmed to alternative RNA targets.
Sequences and putative secondary structures of XNAzyme FR6_1 variants reprogrammed to target (a) human KRAS mRNA residues 426 - 455 (“Sub_KRas84”), (b) human KRAS mRNA residues 677 - 699 (“Sub_KRas166”), (c) human BRAF mRNA residues 2012 - 2038 (“Sub_BRaf600”) and (d) human KRAS residues 213 – 242 (“Sub_KRas12”). Urea-PAGE gels and graphs show bimolecular pre-steady state reactions of the XNAzymes shown (5 uM) with their cognate substrate RNA (1 uM). Incubation times for reactions shown in Urea-PAGE gels were (a & b) 17 h, (c & d) 20 h (25 °C, pH 8.5, 25 mM Mg2+). XNAzymes (c) FR6_1_BRaf600 and (d) FR6_1KRas12 demonstrate the potential for allele-specific targeting, with an obligate requirement for the BRAF c.1799T>A [V600E] or KRAS c.35G>A [G12D] versions of their substrate RNAs, respectively. Data and fits are shown for each re-targeted XNAzyme (black circles and solid black lines) in comparison with the parent FR6_1 cleaving its substrate (Sub_Ebo) under the same conditions (grey squares and dashed grey lines). Error bars show standard error of the mean (s.e.m.) of three independent replicates. All reactions were performed in quasi-physiological conditions (37 °C, pH 7.4, 1 mM Mg2+) unless stated otherwise.