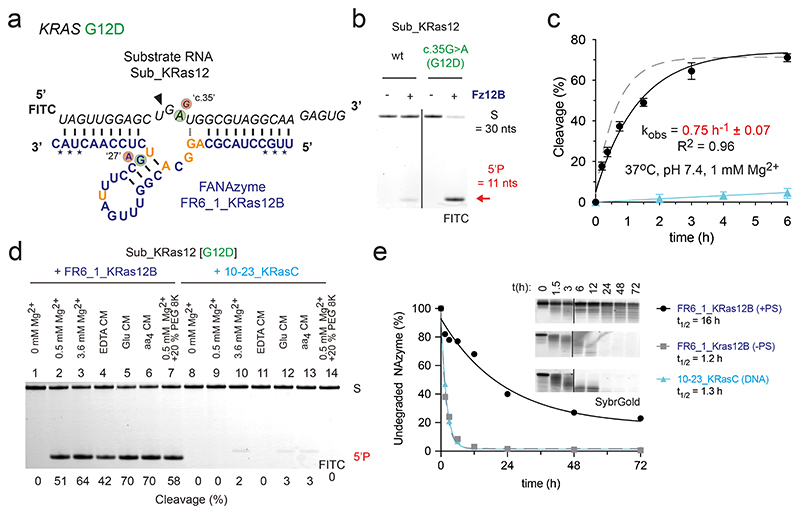
Figure 3. Reselection and modification of an FR6_1 variant yields a biostable allele-specific XNAzyme targeting KRAS G12D RNA.
(a) Sequence and putative secondary structure of XNAzyme “FR6_1_KRas12B” (Fz12B) reselected to target RNA “Sub_KRas12[G12D]”, comprising human KRAS RNA residues 213 – 242 bearing the G12D (c.35G>A) mutation. FANA residues in the catalytic core differing from FR6_1 are shown in gold. FANA-phosphorothioate modifications are denoted by (*). Circled bases denote mutations in the substrate or XNAzyme that enable (green) or abolish (red) RNA cleavage. (b) Urea-PAGE gel (17 h) and (c) graph showing bimolecular pre-steady state reactions of RNA Sub_KRas12 [wild-type (wt), or G12D] (1 uM) with enzymes (5 uM): data and fits are shown for Fz12B (black circles and solid black line) and an analogous DNAzyme “10-23_KRasC” (see Supplementary Fig. 4) (cyan triangles and solid cyan line) cleaving Sub_KRas12 [G12D], and the parent FR6_1 cleaving its cognate RNA substrate (Fig. 1a, 2a) for reference (Dashed grey line). (d) Urea-PAGE gel showing cleavage of Sub_KRas12[G12D] RNA (1 uM) by Fz12B (5 uM) or 10-23_KRasC (5 uM) in buffers mimicking intracellular conditions63 (See Supplementary Table 2)(2 h). Differences in product size are due to 10-23_KRasC cleaving one residue downstream compared with Fz12B. (e) Urea-PAGE gel and graph showing biostability of Fz12B with FANA-phosphorothioate (PS) modifications (black circles) or without (grey squares), and 10-23_KRasC (cyan triangles) in 90% human serum at 37 °C. All cleavage reactions shown were performed in quasi-physiological conditions (37 °C, pH 7.4, 1 mM Mg2+) unless stated otherwise.