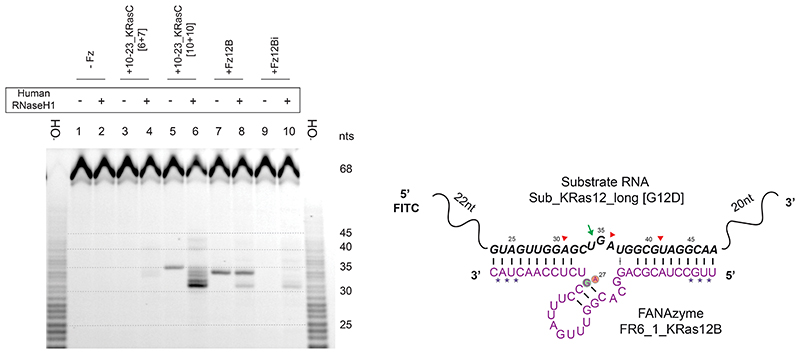
Extended Data Fig. 4. Characterisation of RNase H1-dependent RNA cleavage induced by active and inactive XNAzymes and DNAzymes.
Urea-PAGE gel showing assays of human RNase H1-mediated cleavage of RNA substrate Sub_KRas12_long [G12D], induced by XNAzyme FR6_1_KRas12B (“Fz12B”), catalytically-inactive FR6_1_KRas12B[G27A] (“Fz12Bi”), or DNAzymes 10-23_KRasC [6 + 7] or 10-23_KRasC [10 + 10]. Sequences and putative secondary structures of Fz12B and target RNA show deduced locations of cleavage sites mediated by RNase H (red arrows) or the FANAzyme alone (green arrow). Note that partial alkaline hydrolysis of the RNA substrate (-OH) and XNAzyme-mediated cleavage produce 5’ RNA products that terminate in 3’ cyclic phosphate, whereas RNase H-mediated cleavage produces 3’ OH termini; the resulting difference in PAGE mobility has been taken in account when deducing cleavage sites.