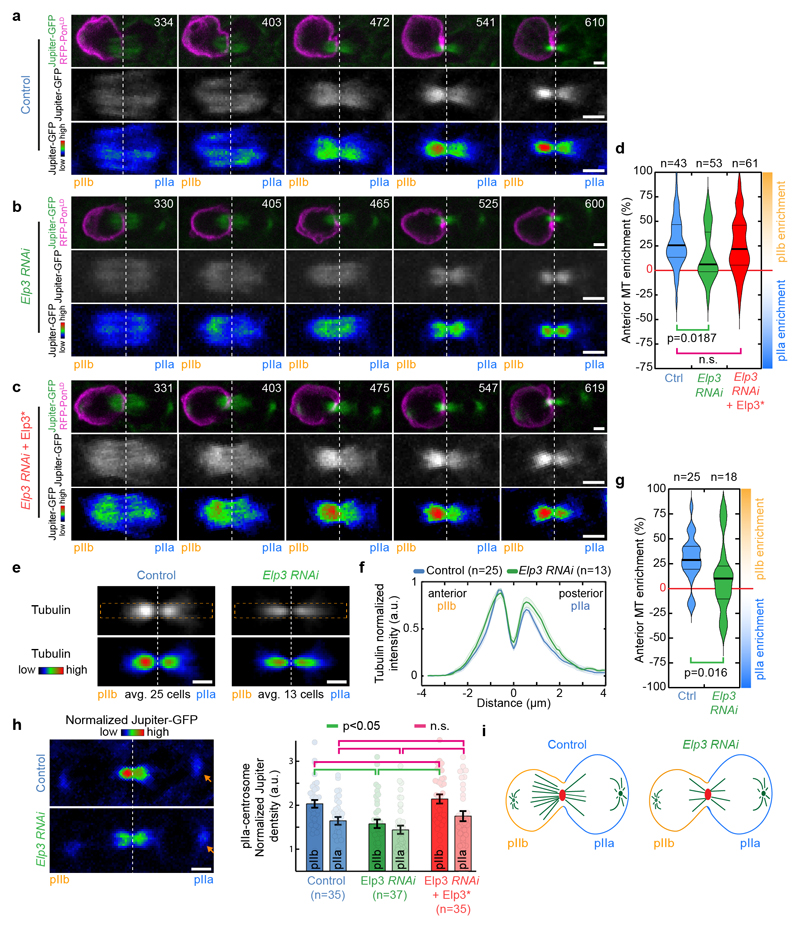
Figure 1. Elongator controls central spindle asymmetry.
(a-c) Timelapse of dividing SOP of indicated genotypes showing Jupiter-GFP/mRFP-PonLD (top rows); Jupiter-GFP (middle) or Jupiter-GFP with Rainbow RGB LUT (bottom rows). Movies were automatically registered in time (see Methods). Registered time=0s indicates anaphaseB onset. Anterior/pIIb orientation determined by the mRFP-PonLD signal. Images correspond to Spinning Disk Confocal Microscopy (SDCM) imaging (maximum intensity projection). (d) Enrichment in pIIb (see methods) for conditions shown in a-c. Thick line: median; thin line: quartile; n: number of SOPs analysed. Statistics: Kruskal-Wallis non-parametric ANOVA1 test (respective post-hoc test P-values indicated). Central spindles in WT SOPs are asymmetric, with an enrichment of Jupiter-GFP on the pIIb side of the spindle, while in Elp3RNAi they are symmetric. The phenotype can be rescued by expression of an RNAi-resistant version of Elp3, (Elp3*). (e-g) Tubulin immunostainings of central spindles in control and Elp3RNAi mutants. (e: top rows). Average central spindle images of SOPs after anti-β-tubulin immunolabeling, see methods. n: number of spindles. (f) Intensity profile of the β-tubulin signals across the spindle (dashed orange rectangle in e) displaying two peaks, one in pIIa, one in pIIb, due to the exclusion of antibodies from the core of the spindle. In control SOPs the pIIb peak is ≈25% higher than that of the pIIa, and this is abolished in Elp3RNAi mutants. Mean±SEM (shaded area). (g) Enrichment in pIIb. Thick line: median; thin line: quartile; n: spindles analysed. Statistics: two-sided Mann-Whitney test (P-values indicated). Central spindle asymmetry is abolished upon Elongator depletion. (h) Jupiter-GFP signal in dividing SOPs (left panel) and quantification (right panel; Mean±SEM) after normalization to pIIa centrosome (arrowhead, see methods). Statistics: ANOVA2 on square root transformed data, followed by a Tukey multiple comparison test (p value indicated). (i) Elp3 RNAi phenotype summary. Scale bars: 2 μm (a-c, h); 1 μm (e). Source numerical data available in source data.