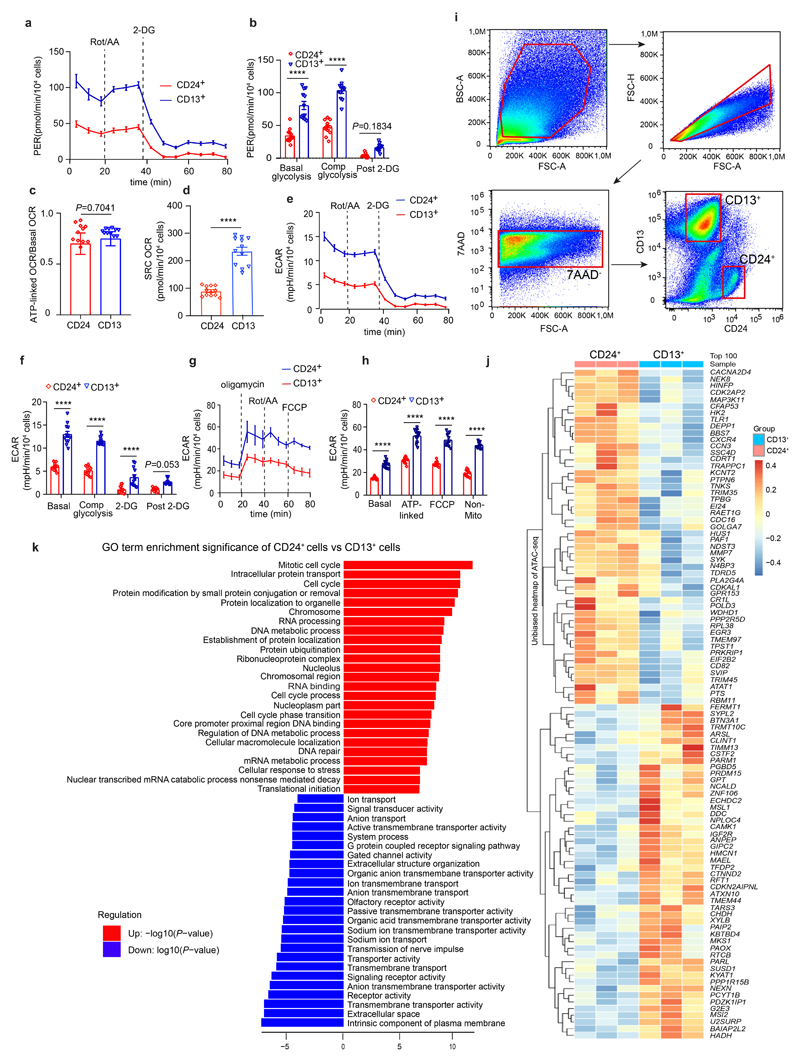
Extended Data Fig. 1. Metabolism and DNA accessibility in CD24+ cells as compared to CD13+ cells.
a-b, Cellular glycolysis and proton efflux rate (PER) of CD24+ cells as compared to CD13+ cells. Statistical analysis in n = 12 mean ± s.e.m. (a-b), 2-way ANOVA post-hoc Bonferroni, non-significant for post 2-deoxy-D-glucose (2-DG, a glucose analog that inhibits glycolysis) in CD24+ cell vs CD13+ cells, P = 0.1834 (b); Basal glycolysis and comp glycolysis (compensatory glycolysis) in CD24+ cells vs CD13+ cells, ****P < 0.0001. c, ATP-linked versus basal oxygen consumption rate (OCR) between CD24+ cells and CD13+ cells in n = 12 mean ± s.e.m., P = 0.7041, non-significant using a two-tailed (also called two-sided) unpaired t-test with Welch’s correction. d, Spare respiratory capacity (SRC) of CD24+ cells compared to CD13+ cells, n = 12 mean ± s.e.m., ****P <0.0001 by a two-tailed unpaired t-test with Welch’s correction. e-f, Extracellular acidification rates (EACR) of CD24+ cells compared to CD13+ cells during anaerobic glycolysis. n = 12 mean ± s.e.m.(e-f), 2-way-ANOVA with post-hoc Bonferroni, non-significant, P = 0.053 for Post 2-DG in CD24+ cells vs CD13+ cells, ****P <0.0001 for Basal, Compensatory glycolysis and 2-DG in CD24+ cells vs CD13+ cells(f). g-h, Extracellular acidification rates (EACR) in CD24+ cells compared to CD13+ cells during aerobic mitochondrial respiration. n = 12 mean ± s.d. (g-h), ****P <0.0001 for Basal, Oligomycin-ATP-linked, FCCP, and non-Mito-Rot/AA in CD24+ cells vs CD13+ cells by 2-way-ANOVA with post-hoc Bonferroni(h). i, FACS gating for sorting of CD13+ or CD24+ cells. j, Scaled heat map showing the chromatin accessibility of top 50 most accessible promoter regions in CD24+ cells or CD13+ cells. k, Top 25 upregulated and downregulated GO terms in CD24+ cells versus CD13+ cells based on the genome-wide measured chromatin accessibility by ATAC-seq.