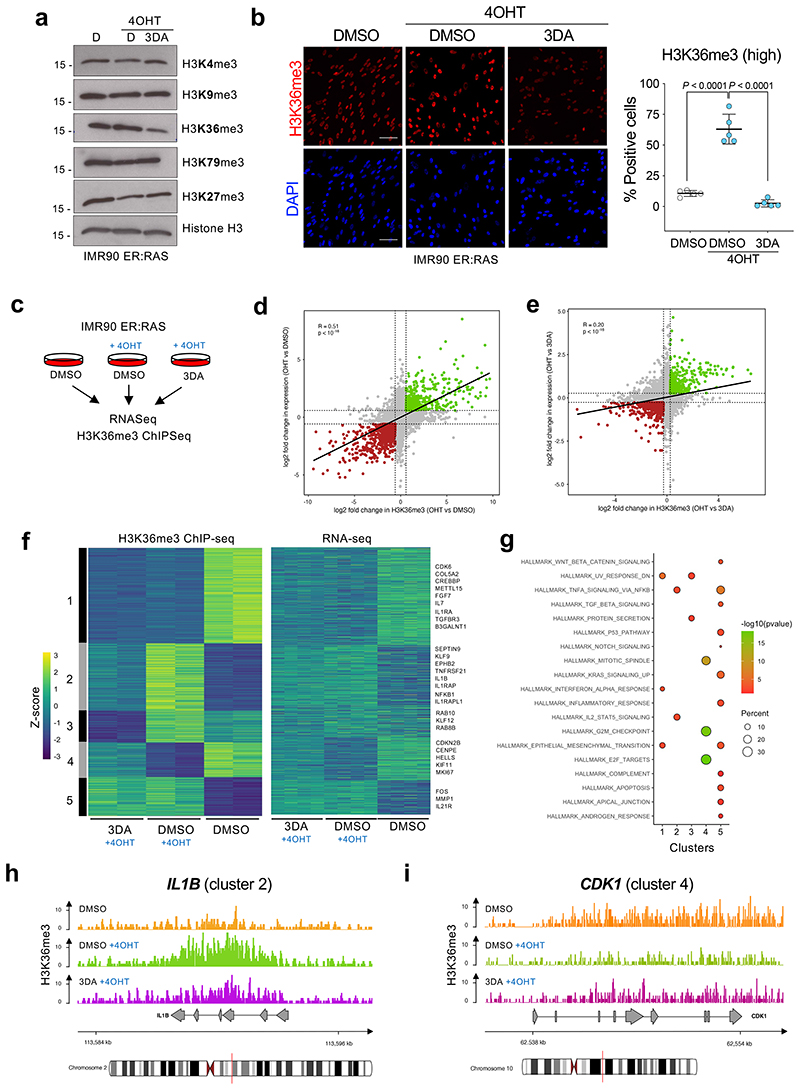
Figure 4. 3DA treatment affects histone H3 K36 methylation during OIS.
a, Immunoblots of histones extracted from IMR90 ER:RAS cells after 4OHT induction and treatment with 10 μM 3DA or vehicle (DMSO). Immunoblot of Histone H3 is included as a sample processing control. Immunoblots are a representative experiment out of three. b, Left, representative immunofluorescence images of H3K36me3 staining (red) in IMR90 ER:RAS cells 4 days after 4OHT induction. Cells were treated with 10 μM 3DA or vehicle (DMSO). Scale bar, 100 μm; right, quantification (n = 5). Statistical significances were calculated using one-way ANOVA. Error bars represent mean ± s.d; n represents independent experiments. c, Experimental design for transcriptional profiling of IMR90 ER:RAS cells 6 days after after 4OHT induction and treatment with 10 μM 3DA or vehicle (DMSO). d, Fold-fold plot comparing fold change in gene expression and ChIP-seq signal in 4OHT versus DMSO treatment. e, Fold-fold plot comparing fold change in gene expression and ChIP-seq signal in 3DA versus 4OHT treatment. The Pearson’s product-moment correlation R and the associated p-value are reported in d and e. f, Heatmap depicting the level of H3K36me3 ChIP-seq signal (Z-score) and the expression level of the closest / overlapping genes (Z-score) for peaks defined as differential between DMSO + OHT vs DMSO, or 3DA + OHT vs DMSO. ChIP-seq data were partitioned into five modules using k-mean clustering. Example genes are displayed on the right side for each module. g, Functional over-representation map depicting enriched hallmark genesets for each module. Circles are colour coded according to the FDR-corrected p-values based on the hypergeometric test. Size is proportional to the percentage of genes in the hallmark gene set belonging to the cluster. hi, Representative genome browser snapshots showing H3K36me3 normalized signal at IL1B (module 2, h) and CDK1 (module 4, i) gene loci for DMSO (orange), DMSO + 4OHT (green) and 3DA + 4OHT (violet) conditions. Data are expressed as normalized counts per million reads (CPM) in 200bp non-overlapping windows.