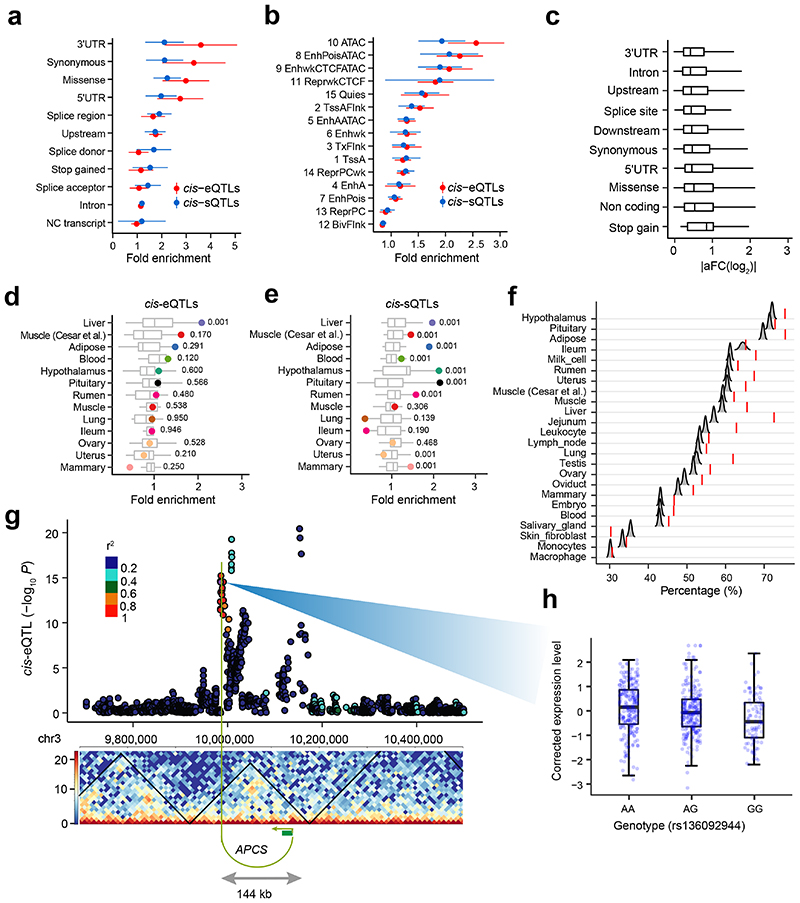
Figure 5. Functional annotation of cis-QTLs.
(a) Enrichment (fold change, the two-sided permutation test with 1,000 times) of cis-eQTLs and cis-sQTLs of 23 distinct tissues in sequence ontology. The data are presented as Mean ± SD. (b) Enrichment (fold change, the two-sided permutation test with 1,000 times) of cis-eQTLs and cis-sQTLs of 23 distinct tissues in 15 chromatin states predicted from cattle rumen epithelial primary cells in Holstein animals15. The data are presented as Mean ± SD. (c) Effect sizes (measured as |aFC(log2|) of cis-eQTLs of 23 distinct tissues across sequence ontology. (d) and (e) Enrichment of cis-eQTLs and cis-sQTLs of 13 tissues in tissue-specific hypomethylated regions, respectively. These 13 tissues have both DNA methylation and cis-QTL data. The numbers are P-values for enrichments of matched tissues (highlighted dots) based on the permutation test (the two sided, 1,000 times). (f) Percentages of eGene-eVariant pairs that are located within topologically associating domains (TADs) are significantly (FDR < 0.01, one-sided) higher than those of random eGene-SNP pairs with matched distances, except for ileum, macrophage and skin fibroblast. The null distributions of percentages of eGene-SNP pairs within TADs are obtained by doing 5,000 bootstraps. The TADs are obtained from the lung Hi-C data. (g) An eGene (APCS) and its eVariant (rs136092944) are located within a TAD, and linked by a significant Hi-C contact (10kb bins, position 9985,000 is linked to 10,135,000 in chr3 with Benjamini-Hochberg corrected P = 1.4×10-6. The P-value is obtained based on the binominal distribution model. The Manhattan plot shows the P-values of all tested SNPs in the cis-eQTL mapping analysis of APCS. The linkage disequilibrium (LD, r2) values between eVariant (rs136092944) and surrounding SNPs are shown in colors. (h) The boxplot shows the PEER-corrected expression levels of APCS across the three genotypes of eVariant (rs136092944), i.e., AA (n = 237), AG (n = 245), and GG (n = 94), respectively.