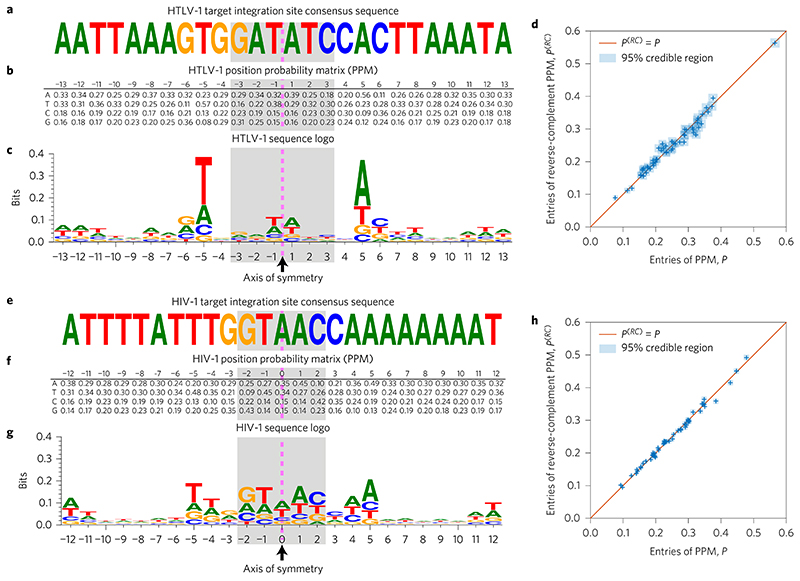
Figure 1. Palindromic HTLV-1 and HIV-1 target integration site consensus sequences and position probability matrices (PPMs), calculated from 4,521 HTLV-1 and 13,442 HIV-1 InS sequences.
a, In agreement with previous studies, we find the HTLV-1 consensus sequence to be a distinctive weak palindrome. The dashed pink line indicates the palindrome’s axis of symmetry, while the shaded area indicates the duplicated region. b, The PPM, P, for the target integration sites is also palindromic; that is, P1,–j ≈ P2,j, P2,–j ≈ P1,j, P3,–j ≈ P4,j and P4-j≈ P3J for j = 1,…,13. Sequence positions to the left of the symmetry line are labelled as negative, and those to the right as positive. c, The symmetry in the PPM may be conveniently visualized using a sequence logo, which also highlights that the palindrome is only weak (has low information content). d, We plot the entries in the first 13 columns of the PPM, P, against the corresponding entries in the reverse-complement PPM, P(RC) (that is, the PPM obtained after first taking the reverse complement of all of the sequences). Uncertainty in the PPM entries is indicated using blue squares showing the 95% credible interval (highest posterior density) range (see Methods). A perfectly palindromic PPM would be one for which P(RC = P, the entries of which would lie along the diagonal shown in the plot. e-h, As in a-d, but using the HIV-1 integration sites.