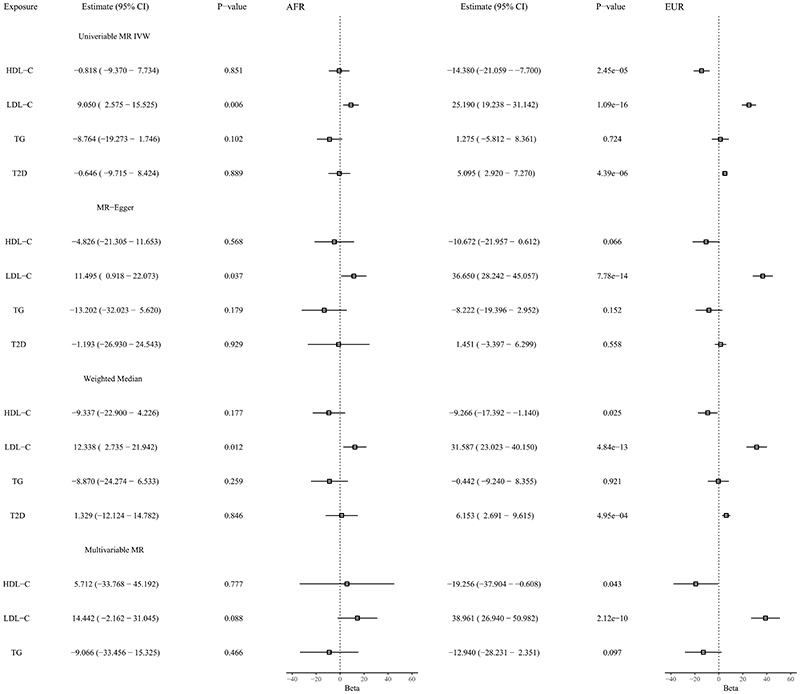
Figure.
Associations between genetically predicted lipid traits, type 2 diabetes mellitus liability, and carotid intima-media thickness from univariable and multivariable Mendelian randomization analyses in African and European ancestry individuals. Estimates (95% CI) represent the estimated increase in carotid intima-media thickness (measured in micrometres) per SD increase in genetically predicted levels of the lipid trait or per unit increase in log odds of type 2 diabetes mellitus. For Africans, the genetic variants explained 13.2% of the variance in LDL-C, 8.4% in HDL-C, and 7.8% in TG. For Europeans, the corresponding values were 7.4% for LDL-C, 9.0% for HDL-C, and 5.5% for TG. The shared SNPs across both ancestries explained 0.18% of the variance in HDL, 0.19% in LDL, and 0.01% in TG for European ancestry individuals, while the corresponding values for Africans were 0.19% for HDL, 0.16% for LDL, and 0.02% for TG. Number of SNPs = 150 (HDL-C), 113 (LDL-C), 133 (TG), and 412 (T2D) in Europeans while in Africans number of SNPs = 55 (HDL-C), 74 (LDL-C), 32 (TG), and 21 (T2D). No proxy SNPs were used. Estimated using univariable MR IVW, MR-Egger, Weighted median and Multivariable MR. Abbreviations: AFR = African, EUR = European, MR = Mendelian randomization, IVW = inverse-variance weighted, HDL-C = high-density lipoprotein cholesterol, LDL-C = low-density lipoprotein cholesterol, TG = triglycerides, T2D = type 2 diabetes mellitus. The genetic variants used in the analyses are available at https://zenodo.org/record/7229645.