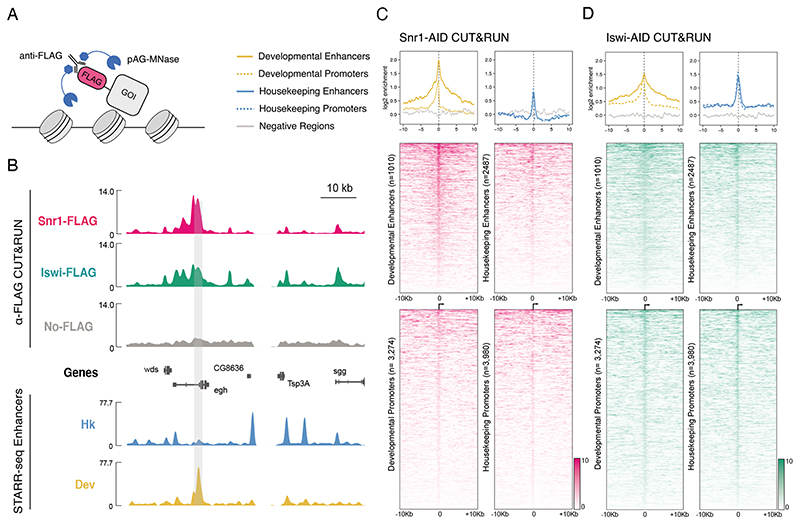
Figure 2. CUT&RUN profiling of Snr1 and Iswi genome occupancy.
A) Depiction of CUT&RUN assay: anti-FLAG antibody is bound to FLAG tagged gene of interest (GOI) and subsequently by AG-MNase fusion protein. B) Chromatin remodeler occupancy in Drosophila S2 cells. Top: genome browser tracks of anti-FLAG CUT&RUN coverage in Snr1-AID cells (pink), Iswi-AID cells (green), and No-FLAG parental cells as control (grey). Bottom: STARR-seq tracks of housekeeping and developmental enhancer activity. A developmental enhancer and the corresponding Snr1 peak is highlighted by grey shading; units: library normalized counts per million. C) Snr1 occupancy at extended genomic regulatory regions (+/- 10kb). CUT&RUN enrichments of FLAG-tagged Snr1 against parental control shown as metaplots (top; colors see legend) and heatmaps (bottom, enrichments as shades of pink, see legend) at developmental (left) and housekeeping (right) enhancers and promoters. D) as (C) but for Iswi (enrichments as shades of green, see legend).