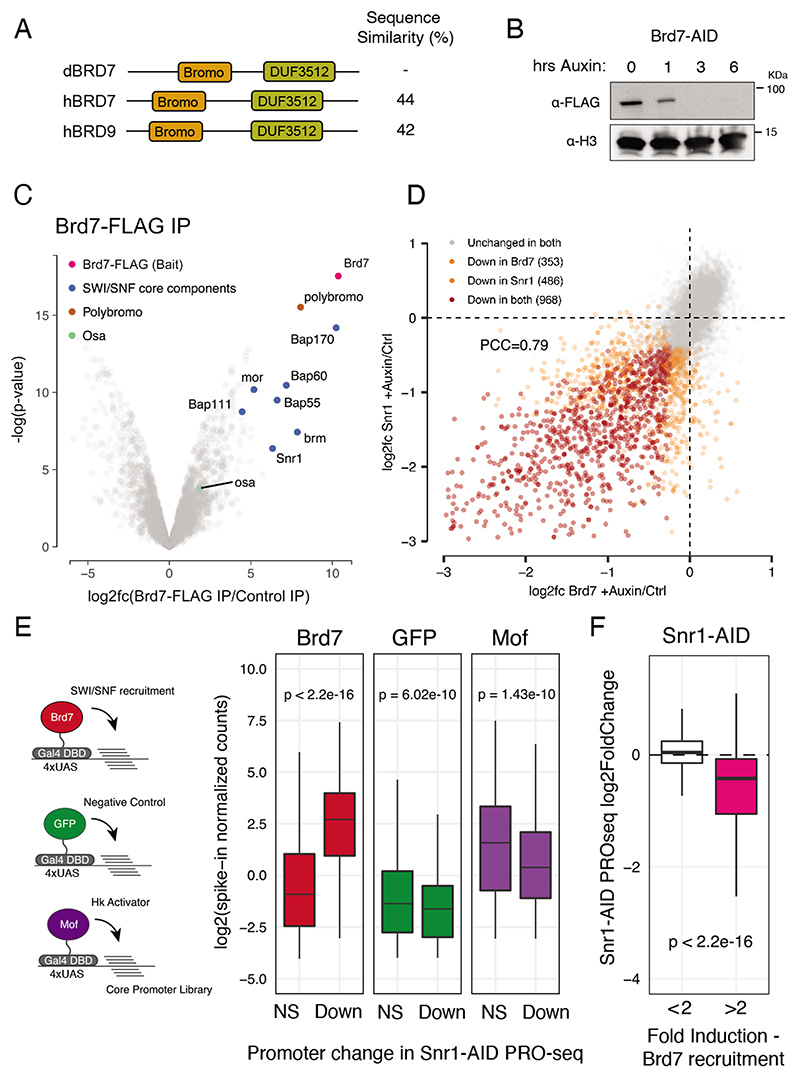
Figure 3. SWI/SNF shows intrinsic activity towards developmental core-promoters.
A) Protein domains of dBrd7 and its human orthologs Brd7 and Brd9. B) anti-FLAG (top) and anti-H3 (bottom) western blot of Brd7-AID S2 cells after 0, 1, 3 and 6 hours of auxin treatment. C) Volcano plot of enrichment and Limma-computed p-value of proteins detected from Brd7-FLAG IP-MS. Highlighted are core components of SWI/SNF in blue and accessory components Polybromo (PBAP specific) and Osa (BAP specific) in in brown and green, respectively. D) Gene expression changes after Snr1 depletion vs. Brd7 depletion.Scatterplot of PRO-seq log2 fold-changes per promoter, with unaffected promoters in grey, uniquely affected promoters in orange, and promoters affected after both depletions in red. E) Assessment of intrinsic core-promoter preference of SWI/SNF. Left: schematic of cofactor-recruitment STAP-seq. Cofactors (colored circles) are recruited to a core-promoter library (lines) via Gal4-DBD. Activation of Snr1 independent (NS) and dependent (Down) promoters as measured by STAP-seq (spike-in normalized counts) after recruitment of Brd7, GFP (negative control) and Mof (housekeeping control). F) Snr1-dependency of promoters activatable by Brd7 recruitment in STAP-seq (pink) versus control (white) shown as PRO-seq changes (log2fc) after Snr1 depletion.