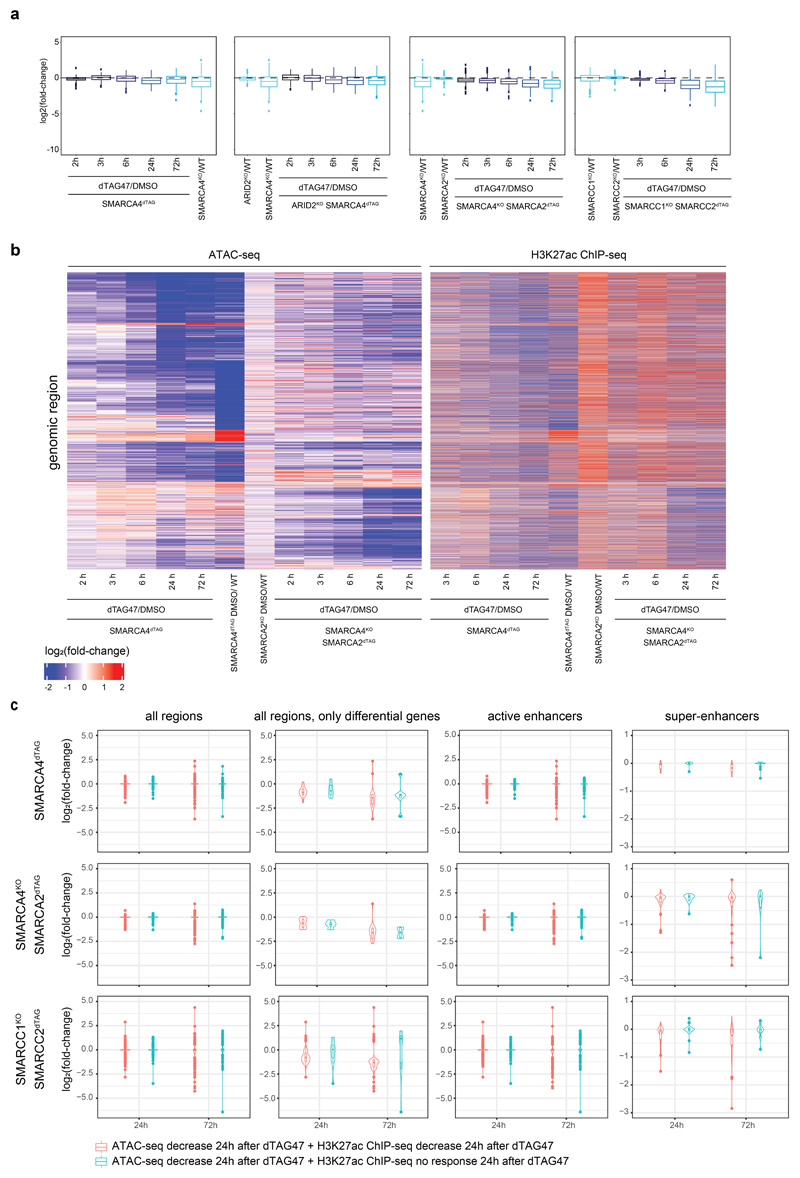
Extended Data Fig. 10. Chromatin and gene-expression alterations after loss of chromatin accessibility at super-enhancers.
(a) Boxplots showing log2 fold-changes of all differential regions annotated as super-enhancers (n=444) over time after dTAG47 treatment in the different HAP1 dTAG cell lines. First and third quartiles are denoted by lower and upper hinges, center is median. The upper/lower whisker extendes to the largest/ smallest value no further than 1.5* inter-quartile range. Data points beyond are plotted individually. (b) Heatmap of log2 fold-changes of ATAC and H3K27ac ChIP-seq signal in WT SMARCA4dTAG and SMARCA4KO SMARCA2dTAG cells after dTAG47 treatment versus DMSO control for the genomic regions differentially accessible in any cell line after TAG47 treatment (cluster 1 – 11). (c) Violin plots of log2 expression fold-changes from RNA-seq experiments for all regions, active enhancer and super enhancer regions showing a decrease in accessibility ATAC-seq signal and either a decrease (log2 fold-change < 1) in H3K27 acetylation ChIP-signal at 24h (red) or no response (log2 fold-change > -1 and < 1) in H3K27 acetylation ChIP-signal at 24h (blue) in SMARCA4dTAG, SMARCA4KOSMARCA2dTAG and SMARCC1KOSMARCC2dTAG cells after dTAG47 treatment (n=2 independent experiments). First and third quartiles are denoted by lower and upper hinges, center is median. The upper/lower whisker extendes to the largest/ smallest value no further than 1.5* inter-quartile range. Data points beyond are plotted individually. Differential genes were defined as padjust < 0.01 und log2(fold-change) > 1.