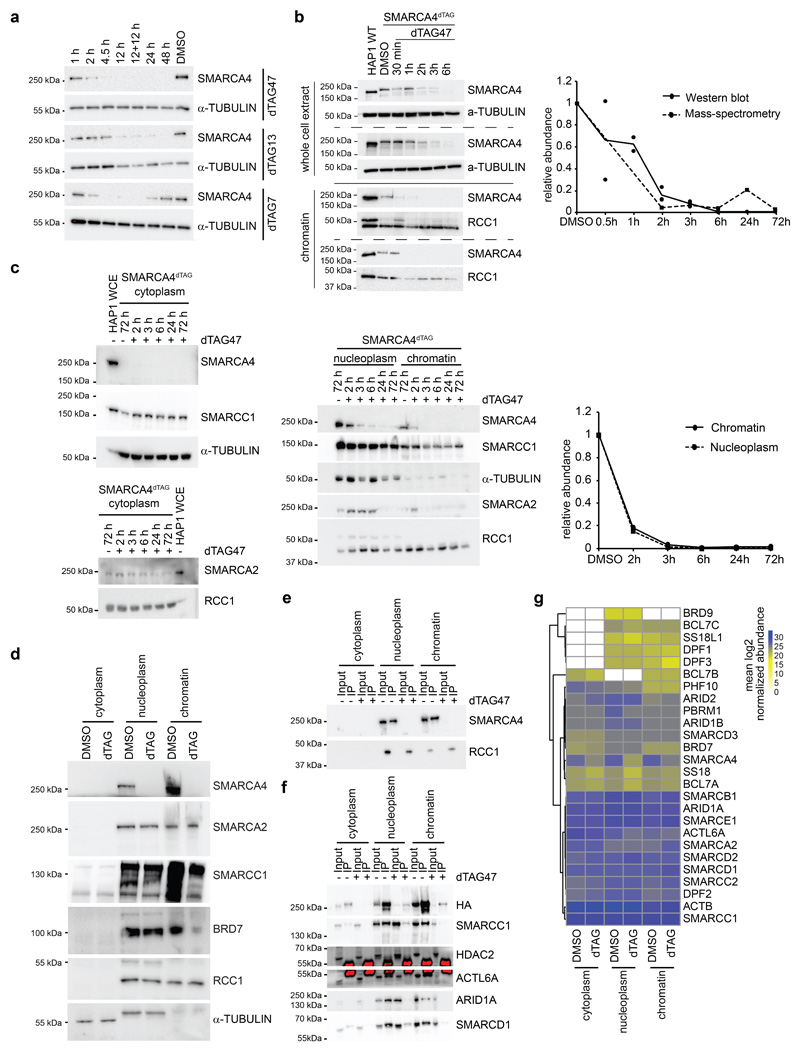
Extended Data Fig. 1. Effects of SMARCA4 degradation on BAF complex members.
(a) Western blot analysis of HAP1 ARID2KOSMARCA4dTAG cells after treatment for different times with 700 nM dTAG47, dTAG13, dTAG7 or DMSO as control (cropped images). (b) Semi-quantitative analysis of SMARCA4 protein levels based on whole-cell extract Western blot data normalized to tubulin (shown in Fig. 1b) and based on nuclear-extract proteomics data (shown in Fig. 1d). Cropped Western blot images of dTAG47 time-course in SMARCA4dTAG cells. (c) Western blot analysis of HAP1 SMARCA4dTAG cells after treatment for different times with 300 nM dTAG47 or DMSO as control in various cellular fractions. Staining for SMARCA4, SMARCA2 and SMARCC1 as well as α-TUBULIN and RCC1 as control. Cropped images, WCE = whole cell extract. Semi-quantitative analysis of SMARCA4 protein levels based on nucleoplasm and chromatin fraction Western blot data normalized to RCC1. RCC1 run on a separate plot from the same experiment and was processed in parallel. (d) Western blot analyses of BAF subunit members after SMARCA4 degradation (dTAG) induced with 300 nM dTAG47 for 24h compared to control (DMSO) in different cellular compartments of HAP1 SMARCA4dTAG cells (cropped images). (e) SMARCC1 immunoprecipitation in different cellular compartments of HAP1 SMARCA4dTAG cells treated with 300 nM dTAG47 or DMSO for 24 h. Western blot analysis for SMARCA4 (cropped images). (f) HA immunoprecipitation in different cellular compartments of HAP1 ARID2KOSMARCA4dTAG cells treated with 300 nM dTAG47 or DMSO for 24 h. Western blot analysis for various BAF subunits (cropped images). (g) Heatmap showing mean log2 SMARCC1-normalized abundance values of mass spectrometry results of figure 1f.