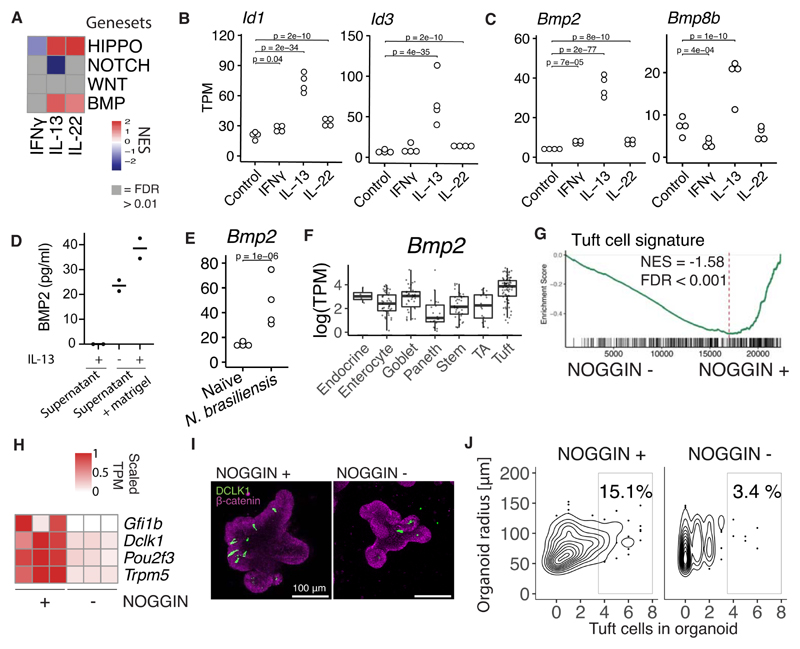
Figure 3. IL-13 induce BMP2 in intestinal epithelium.
A, GSEA analysis of gene sets representing developmental pathways important in IEC development of bulk RNAseq data from organoids treated with 10 ng/mL cytokine for 24 hours. Gene set sources: HIPPO: Genes up in artifical YAP induction and down-regulated in YAP-KO. WNT: Genes up in organoids treated with the GSK3 inhibitor CHIR, NOTCH: Genes up in organoids from ATOH1 KO epithelium compared to wt and BMP: Genes up in organoid treated with BMP4 compared to control. Gene sets and references in supplementary file 1. B,C, Gene expression in small intestinal organoids treated with cytokines for 24 hours determined with RNAseq. Each dot is one biological replicate. D, Concentration of BMP2 in supernatant or supernatant+matrigel (without organoids) of organoids stimulated with IL-13 determined with ELISA. Each dot is one biological replicate. Representative of two experiments. E, Gene expression in epithelium from duodenum extracted from mice infected with N. brasiliensis determined with RNAseq. Each dot is one biological replicate. F, Plate based scRNAseq expression data from Haber et al in small intestinal epithelium. TA = Transit amplifying. G, GSEA of tuft cell gene set on RNAseq of small intestinal organoids grown in the presence of NOGGIN or not for 5 days since splitting. H, Tuft cell marker genes from same RNAseq data as G. I, DCLK1 confocal staining of small intestinal organoids grown in the presence of NOGGIN or not for 72 hours since splitting. Images are projections of Z-stacks. J, Quantitation of experiment in I. Representative of two experiments. Plot shows a total of 462 organoids. TPM = transcripts per million, NES = normalized enrichment score.