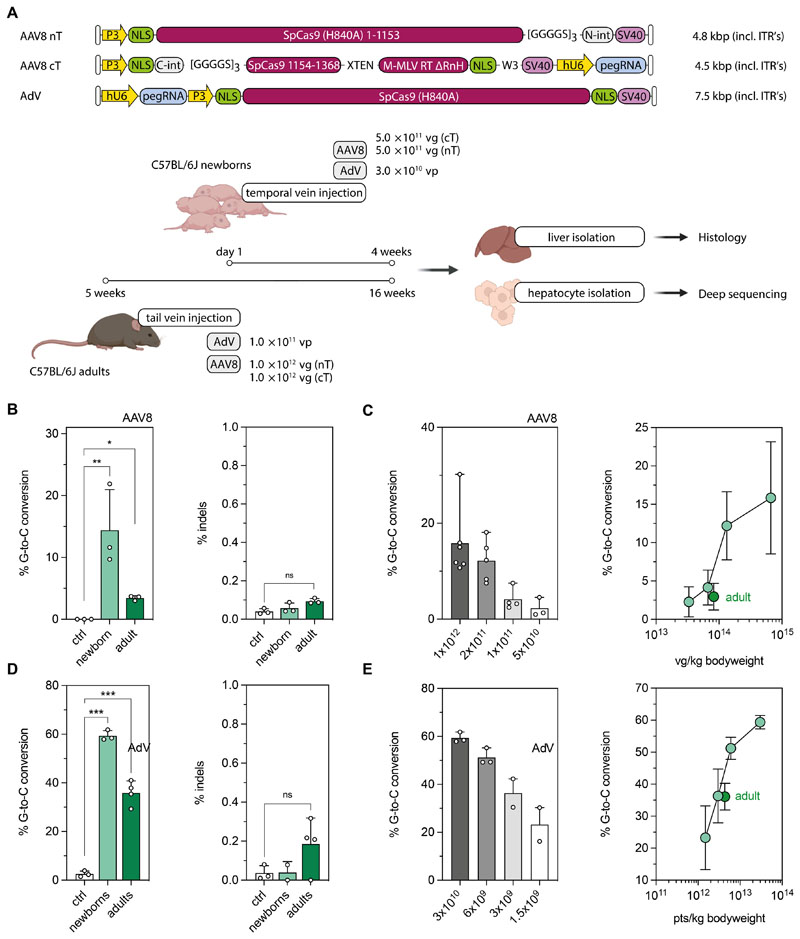
Figure 2. AAV8- and AdV-mediated prime editing at the Dnmt1 locus in the mouse liver.
(A) Schematic outline of the experimental setup with AAV8- or AdV-mediated prime editing in newborn and adult mice. Constructs used for in vivo prime editing at the Dnmt1 locus in the mouse liver are not depicted to scale. (B) Correction and indel rates in newborn and adult animals after AAV8-mediated delivery (injected dose per neonate and adult: 1 × 1012 and 2× 1012 vector genomes; vg). Untreated mice were used as negative controls. Percentage of sequencing reads with indels around the protospacer region were determined by deep sequencing. (C) Editing rates in neonates relative to AAV doses per animal (left panel) and per kg bodyweight (right panel). The respective values for adult mice were added (dark green) for comparison. (D) Correction and indel rates in newborn and adult mice at 4 weeks after AdV-mediated delivery (injected dose per neonate and adult: 3×1010 and 1×1011 viral particles, vp). (E) Editing rates in neonates relative to AdV doses per animal (left panel) and per kg bodyweight (right panel). The respective values for adult mice were added (dark green) for comparison. Untreated mice were used as negative controls. Data are represented as mean ± s.d. (n=3-6 mice per group) and were analyzed using a two-way ANOVA with Tukey’s multiple comparisons test (ns, not significant, P>0.05; *P<0.05; **P<0.005).