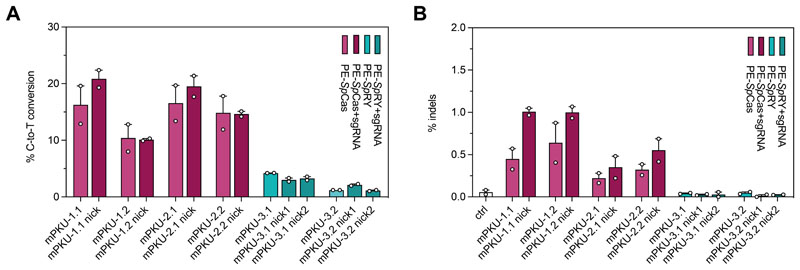
Figure 3. In vitro correction of the Pahenu2 allele using PE-SpCas and -SpRY variants.
(A, B). In vitro editing rates (A) and indel formation (B) of pegRNAs designed for SpCas-(light and dark purple) and SpRY-PEs (light and dark green) to target the disease-causing Pahenu2 mutation (c.835T>C; p.F263S) on exon 7. pegRNAs for the SpCas-PE were also combined with an additional PE3 nicking sgRNA (pegRNA+sgRNA, dark purple). Two nicking sgRNAs were designed for two PE3b approaches using the SpRY variant (dark green). Experiments were performed in reporter HEK293T cells in which the mutated exon 7 of the Pahenu2 gene was stably integrated. Data are represented as mean ± range of two independent experiments.