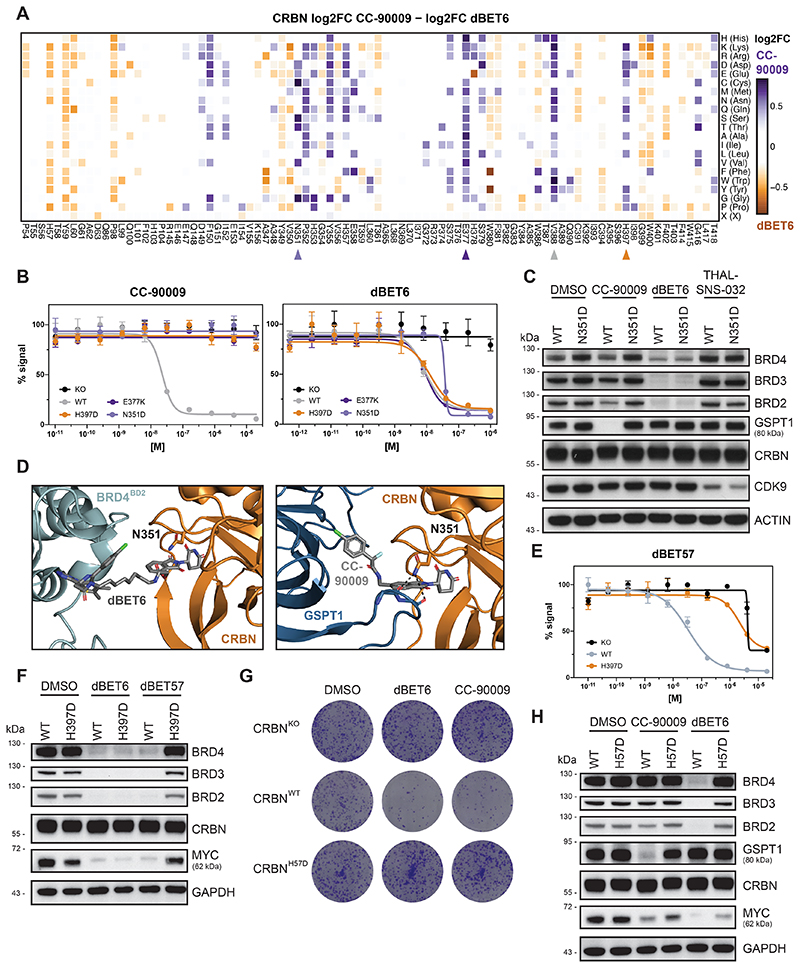
Figure 5. Functional CRBN Hotspots Show Degrader Selectivity and are Mutated in Refractory Multiple Myeloma Patients.
(A) Heatmap depicting differential log2 fold-enrichment of CRBN mutations normalized to maximum log2 fold-changes vs. DMSO between BET bromodomain targeting PROTAC dBET6 (500 nM, 7 d treatment) and the GSPT1 targeting molecular glue CC-90009 (500 nM, 7 d treatment). n = 3 independent measurements.
(B) Dose-resolved, normalized viability after 3 d treatment with CC-90009 and dBET6 in RKO CRBN-/- cells with over-expression of CRBNWT, CRBNE377K, CRBNN351D and CRBNH397D. Mean ± s.e.m.; n = 3 independent treatments.
(C, F and H) Protein levels in RKO CRBN-/- cells with over-expression of CRBNWT, CRBNN351D, CRBNH397D or CRBNH57D treated with DMSO, CC-90009 (50 nM, 6 h), dBET6 (15 nM, 2 h), dBET57 (240 nM, 2 h) or THAL-SNS-032 (200 nM, 2 h). Representative
images of n = 2 independent measurements.
(D) Cocrystal structure of dBET6 (left) and CC-90009 (right) in a ternary complex with CRBN and BRD4BD2 (PDB 6BOY) or GSPT1 (PDB 6XK9) depicting PPIs of CRBNN351 and the GSPT1.
(E) Dose-resolved, normalized viability after 3 d treatment with dBET57 in RKO CRBN-/- cells with over-expression of CRBNWT and CRBNH397D. Mean ± s.e.m.; n = 3 independent treatments.
(G) Depiction of clonogenic assays via crystal violet staining. Cells were treated for 10 days at EC90 of the degrader (30 nM dBET6, 60 nM CC-90009). Representative of n = 2 independent measurements.
See also Extended Data Fig. 6.