Extended Data Figure 1.
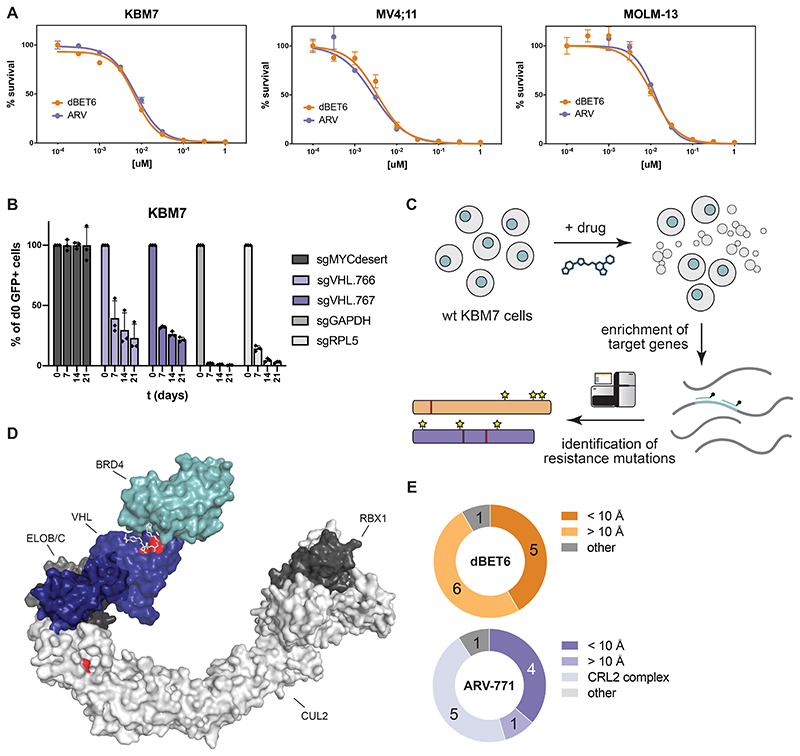
(A) Dose-resolved, normalized viability after 3 d treatment (dBET6 or ARV-771) in KBM7, MV4;11 and MOLM-13 cells. Mean ± s.e.m.; n = 3 independent treatments.
(B) Histogram depicting growth competition experiments. WT control KBM7 cells were mixed with mCherry and Cas9 expressing KBM7 cells harboring sgRNAs against the indicated genes. Pools were flow cytometry quantified at days 0, 7, 14 and 21 and mCherry percentages were normalized to day 0 percentage and to a non-targeting control sgRNA (sgMYCdesert). Data represents mean ± s.d. of n = 3 biological replicates.
(C) Scheme of targeted hybrid-capture approach coupled to next-generation sequencing to identify mutations in spontaneously resistant cells.
(D) Structure depiction of the CUL2-VBC-MZ1-BRD4 complex (PDBs: 5N4W, 5T35). Residues marked in red were identified in hybrid capture analysis. See also Figure 1 and Supplementary Dataset.
(E) Number of spontaneous degrader resistance alterations in the substrate receptor (CRBN, VHL, colored) binned by their distance to the degrader binding site. See also Figure 1D and Supplementary Dataset.