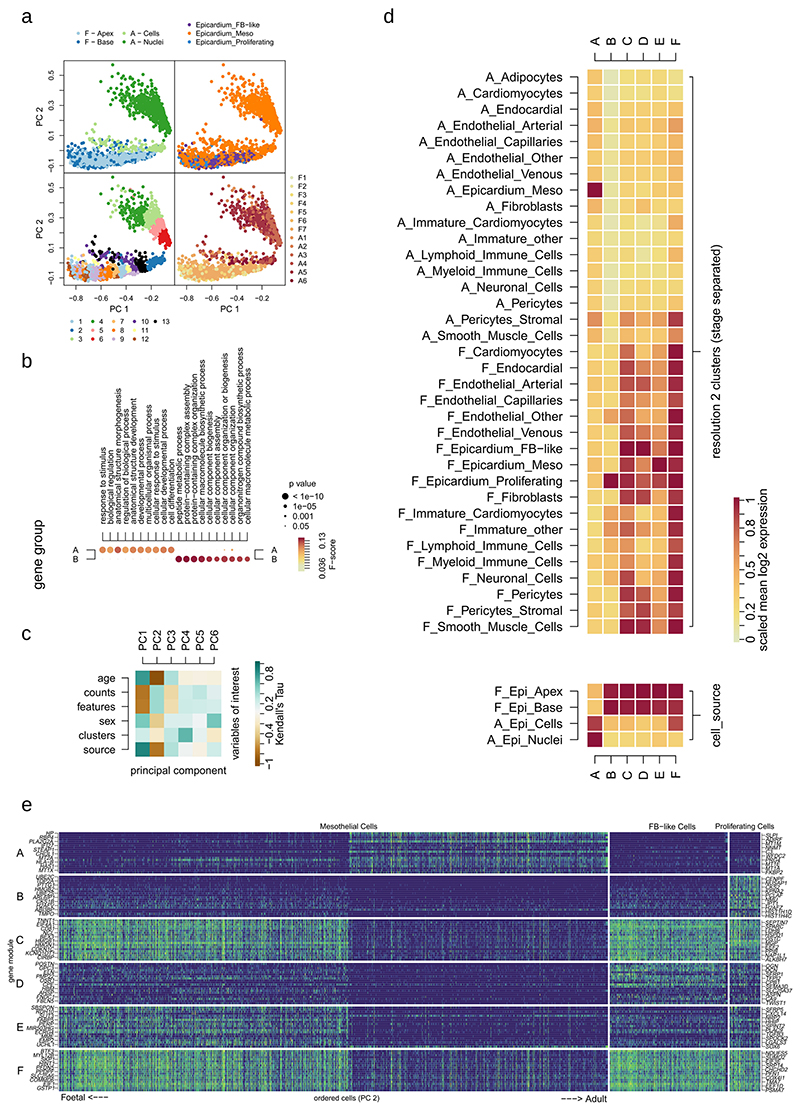
Extended Data Fig. 3. Further observations and technical hurdles in generating epicardial gene modules.
The gene expression matrix of epicardial cells was isolated from the rest and binarised. Co-occurrence clustering of genes including nuclei in the matrix created two gene modules and a, principal component analysis (PCA) on cellular commitment to modules confirms that the main separation of the dataset was between cells and nuclei. Gene Ontology enrichment was carried out; b, revealing broad terms associated with nuclear or cytosolic compartments. Use of Kendall’s rank correlation; c, between cell variables and PCA components with components 2 and 4 highly correlating with age and clusters resepctively as variables of interest for visualisation. d, the mean expression of the 6 gene modules in the 19 resolution-2 clusters as well as cells grouped by source and; e, the expression of the top 20 genes in each gene module across age-ordered epicardial cells (age component PC2).