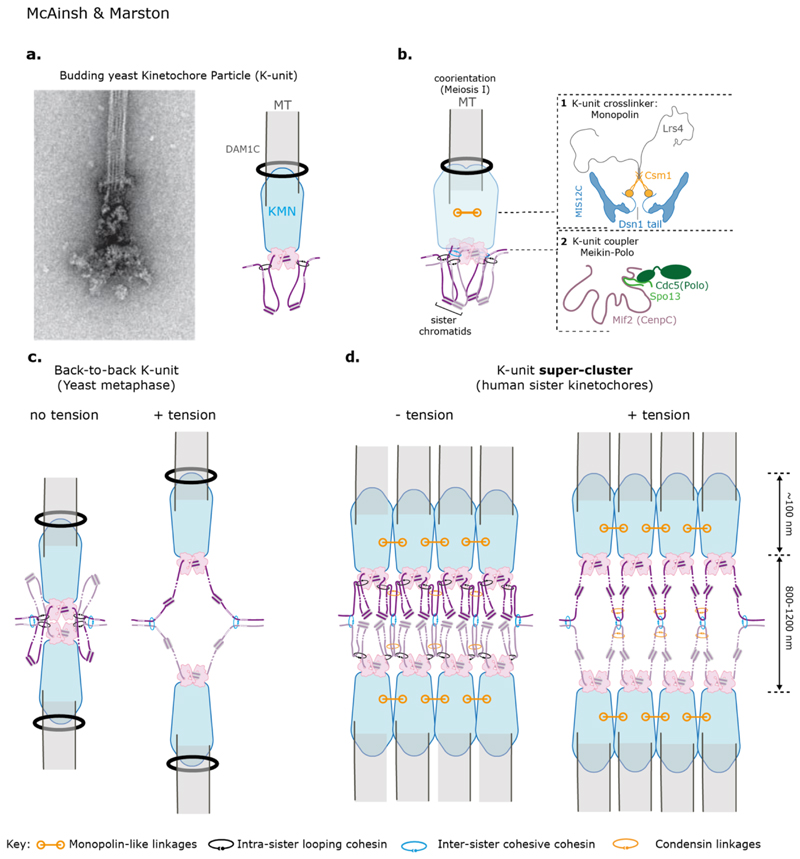
Figure 3. Super-cluster for centromere-kinetochore multimerisation.
(a) left Cryo-electron microscopy image of isolated budding yeast kinetochore particles bound to microtubules. In the image, globular domains contact the microtubule which is encircled by a ring-like structure, likely DAM1C. There is also a central hub which does not contact the microtubule directly. Right Model for the architecture of a single k-unit where one kinetochore superassembly contacts one microtubule, as in budding yeast. Each CCAN anchors cohesin, which forms intramolecular loops on each side of the kinetochore.
(b) Model for sister kinetochore coorientation during meiosis I in budding yeast. Two k-units - the sister kinetochores - are clamped together in a side-by-side orientation due to two kinds of linkages. (1) Monopolin binds to the Dsn1 subunit of the MIS12C and fuses the sister kinetochore together. (2) Meikin-Polo associates with CenpC/Mif2 and promotes coorientation, possibly by facilitating cohesin-dependent-linkages of sister centromeres. Note that a fused pair of sister kinetochores binds a single microtubule in meiosis I (261).
(c) Schematic showing the architecture of the budding yeast kinetochore in mitosis in the presence and absence of spindle tension. Left: CCAN-anchored cohesin extrudes a loop on either side of the centromere until blocked by convergent genes at pericentromere borders. Borders also retain inter-sister, cohesive cohesin. This state, as shown, is short-lived because the attachment of sister kinetochores to microtubules from opposite poles results in the generation of tension. Right Under tension, the chromatin loops extend into a V-shaped structure. Intra-molecular, loop-extruding cohesin slides off but inter-molecular, cohesive, cohesin is trapped at the borders and holds the sister chromatids together.
(d) Speculative model for the architecture of the mammalian kinetochore, inspired by the structure of the budding yeast kinetochore and pericentromeric chromatin in mitosis and meiosis. Ordered arrays of k-units are clustered together. This clustering is facilitated by cohesin anchored on CCAN and stabilised by cross-linkers between KMN, analogus to monopolin. Chromatin-organising complexes such as condensin may further serve to stabilize interactions between adjacent K-units.