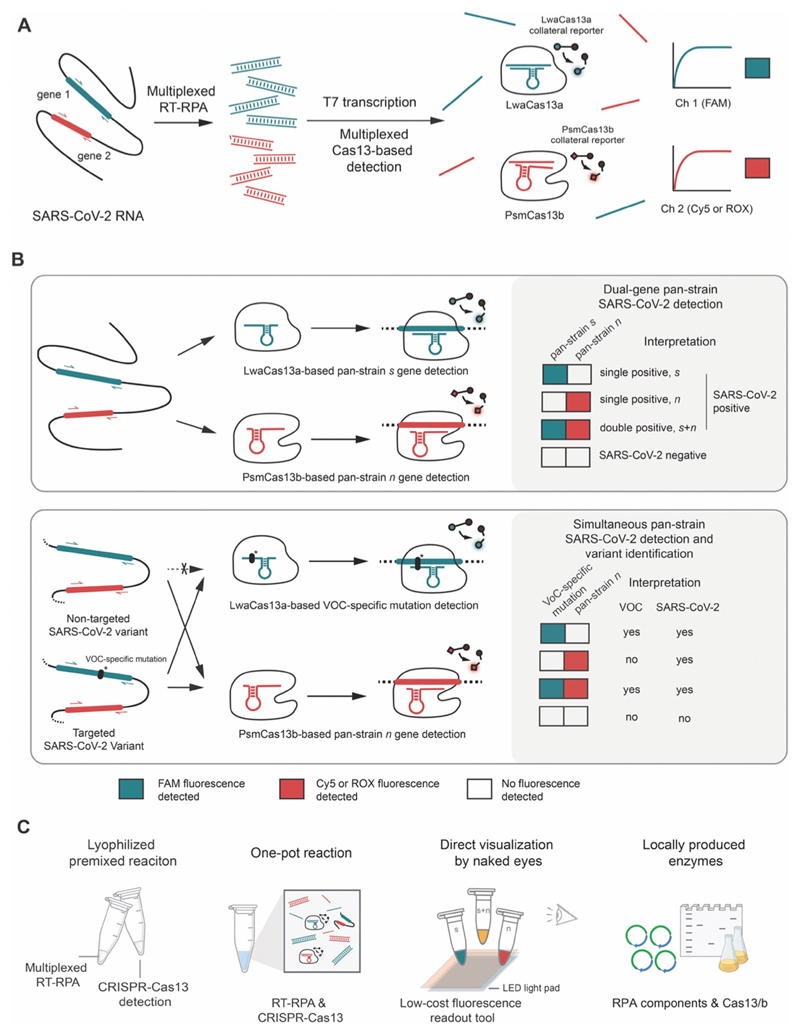
Figure 1. A multiplexed Cas13-based assay for simultaneous SARS-CoV-2 detection and variant identification.
(A) Regions of interest in SARS-CoV-2 RNA, such as the s and n genes, are isothermally amplified by multiplexed RT-RPA. T7 transcription then converts and amplifies DNA amplicons to RNAs, which are recognized by cognate Cas13a-crRNA and Cas13b-crRNA complexes with orthogonal collateral cleavage preferences. Cleavage of orthogonal RNA reporters by target-activated Cas enzymes elicits multicolored fluorescence, which can be monitored with standard fluorescence detection instruments.
(B) Top, dual-gene detection via CRISPR-Cas13a/b for COVID-19 diagnosis. Bottom, dual-gene detection for simultaneous COVID-19 diagnosis and SARS-CoV-2 variant identification. LwaCas13a is used to detect a universal, pan-strain region (top) or a variant-specific region (bottom) of the SARS-CoV-2 s gene. PsmCas13b is used to detect a pan-strain region of the n gene. The amplified regions of the S and N genes are shown in red and green respectively.
(C) Developing simple-to-use SARS-CoV-2 variant surveillance tools which can be locally manufactured. Our multiplexed CRISPR-based detection reactions can be premixed and lyophilized for convenient use and storage; multiplexed RPA and Cas13-based reactions can be combined in one tube; result readouts can be directly observed by eye with low-cost light sources; and RPA and Cas13a/b can be locally produced and formulated.