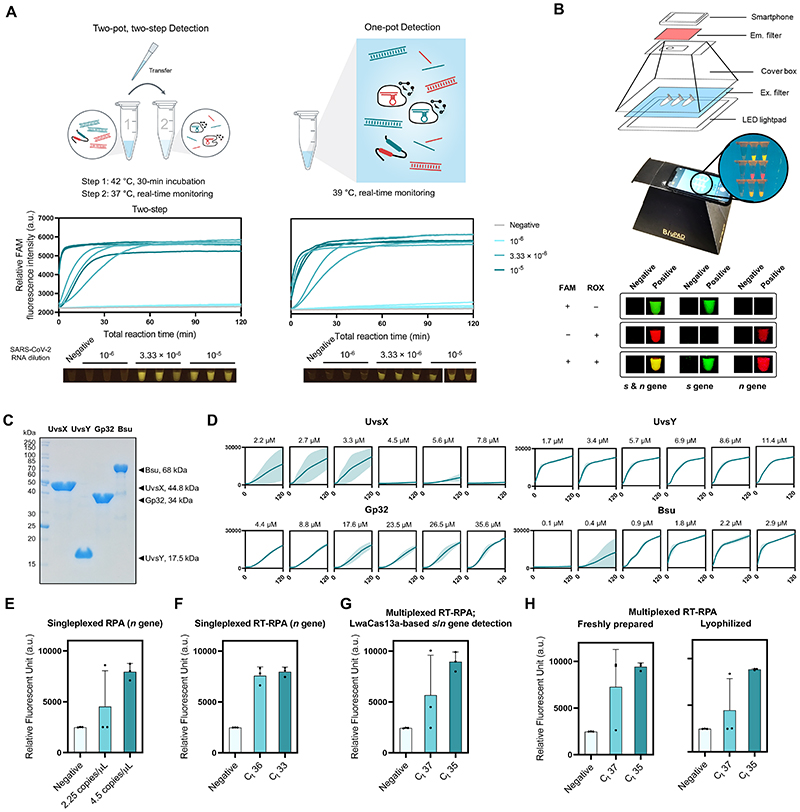
Figure 4. One-pot formulation, easy visualization, and local production of components for RPA and multiplexed CRISPR-based detection.
(A) The one-pot reaction scheme and its analytical sensitivity compared to the standard two-pot setup. For all, s- and n-gene amplicons generated from multiplexed RPA were detected with LwaCas13a-based reaction programmed with s- and n-targeted crRNAs. Data from 3 independent replicates at each RNA input dilution are shown. (B) Easy visualization of multiplexed Cas13-based detection. A sample equipment setup with a LED transilluminator, appropriate lighting gels for visualization of fluorescein (FAM) and rhodamine X (ROX), and a smartphone for image capturing is shown. Smartphone-captured images of FAM and ROX signal generated from multiplexed Cas13a/b-based detection of the s and n genes of SARS-CoV-2. (C) SDS-PAGE analysis of locally produced and purified RPA components: UvsX, UvsY, Gp32, and Bsu polymerase large fragment (Bsu LF). (D) Optimizations of protein concentrations in in-house RPA. RPA reactions with varying concentrations of UvsX, UvsY, Gp32, and Bsu LF were performed with pUC57-2019-nCoV-N plasmid as a DNA template (at 10,000 copies/μl). Thereafter, the RPA products were used in a LwaCas13a-based detection. FAM fluorescence generated over 120 min for each RPA condition is shown. (E, F) Optimally formulated in-house RPA in amplification of the n gene of SARS-CoV-2 from pUC57-2019-nCoV-N plasmid (E) and serially diluted SARS-CoV-2 RNA (F). LwaCas13a-based detection was then used to detect n-gene amplicons, with FAM fluorescence intensity generated after 90 min at 37°C shown. (G) Optimized in-house RT-RPA for multiplexed detection. Multiplexed RT-RPA to amplify the s and n genes of SARS-CoV-2 was performed using serially diluted SARS-CoV-2 RNA as a template. Detection of s and n amplicons was monitored through FAM fluorescence signal generated by Cas13a-mediated s and n gene detection. The reaction was measured after 90 min at 37°C. (H) Lyophilized multiplexed in-house RT-RPA has similar sensitivity as freshly prepared reactions. Amplification and detection of the SARS-CoV-2 s and n gene are as in (G). Data are mean ± s.d. from 3 replicates. RNase-free water was used as input of all negative control reactions.