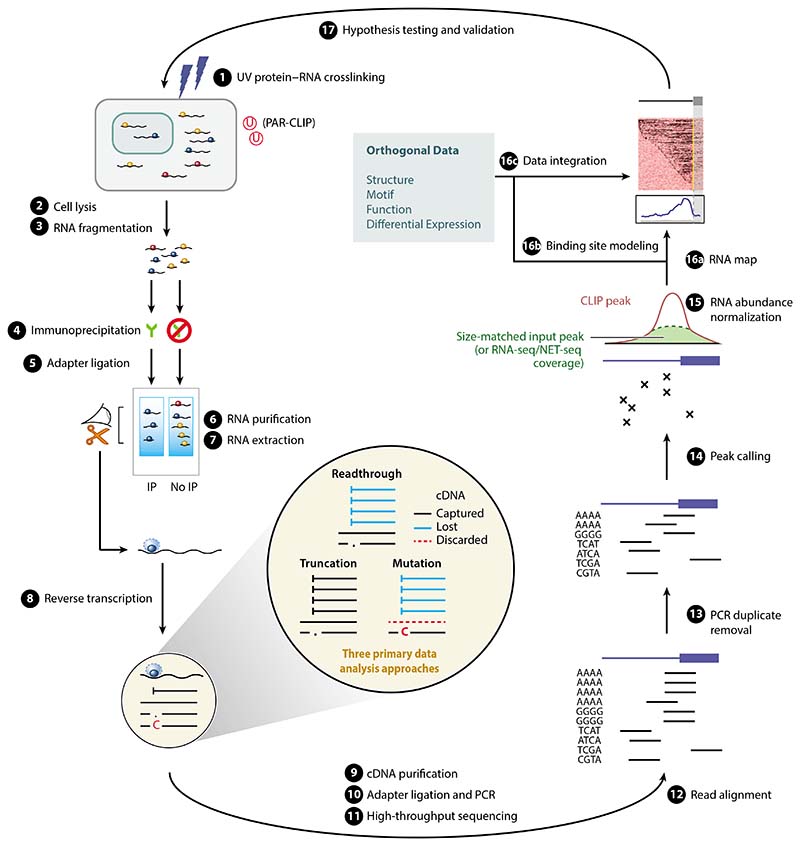
Figure 1.
A computational biologist’s overview of the CLIP method, and its key experimental (left) and computational (right) steps. The experimental steps, common across most methods, are numbered according to the scheme in Reference 7. Highlighted in the center are the three primary data analysis approaches that rely on cDNA readthrough, mutation, or truncation, depending on the type of CLIP protocol used to generate the data (related to Table 1). The cDNAs that are captured by representative protocols are in black, while those that are lost during reverse transcription are in blue, and those that are discarded during analysis are marked by dashed lines. Abbreviations: cDNA, complementary DNA; CLIP, UV crosslinking and immunoprecipitation; IP, immunoprecipitation; NET-seq, native elongating transcript sequencing; PAR-CLIP, photoactivatable ribonucleoside–enhanced CLIP; PCR, polymerase chain reaction; RNA-seq, RNA sequencing.