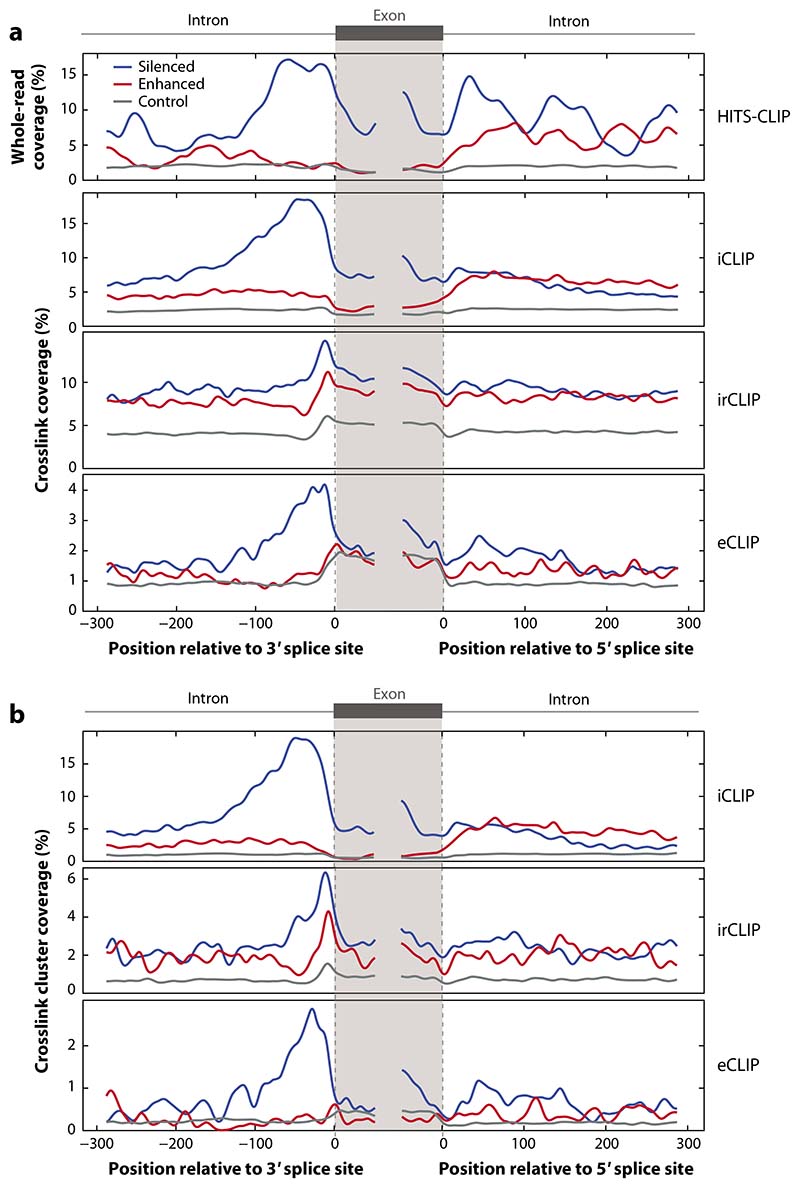
Figure 3.
Using RNA maps to examine sensitivity and specificity of CLIP data. PTBP1 is an abundant RBP that crosslinks efficiently and follows a position-dependent regulatory mechanism, thus making it suitable for data analysis via an RNA map. The regulated exons were defined as those with an absolute dIrank of greater than 1 on analysis of splice junction microarray data with ASPIRE3 software upon knockdown of PTBP1/PTBP2 in HeLa cells (99). In panel a, we compare the raw data for different experimental methods, with whole reads from HITS-CLIP in HeLa cells (100), crosslink positions from irCLIP (18) and iCLIP (16) in HeLa cells, and eCLIP in HepG2 cells (25). This demonstrates that CLIP data can lead to strong enrichments even without peak calling, but this depends on the specificity of data. In panel b, we analyze the effects of peak calling on the crosslink positions from different experiments, with data from irCLIP (18) and iCLIP (16) in HeLa cells and eCLIP in HepG2 cells (25) analyzed using the iCount peak caller with 15 nucleotide clustering (15). The code to reproduce this figure is available for download at https://github.com/jernejule/clip-data-science. Abbreviations: CLIP, UV crosslinking and immunoprecipitation; eCLIP, enhanced CLIP; HITS, high-throughput sequencing; iCLIP, individual-nucleotide resolution CLIP; irCLIP, infrared CLIP; RBP, RNA-binding protein.