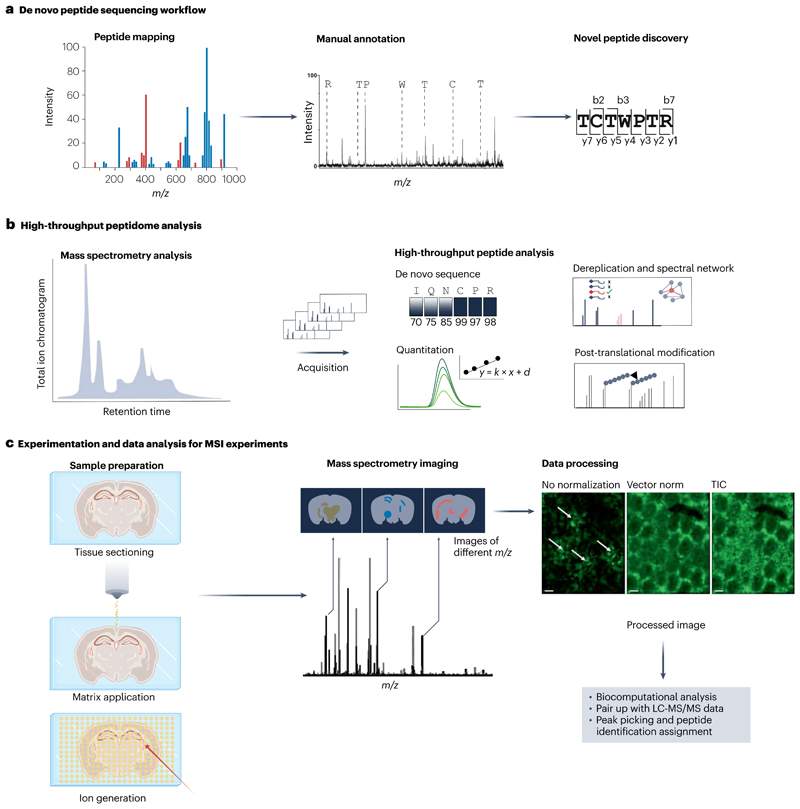
Fig. 3. Overview of common peptidomics workflows.
a, De novo peptide sequencing is utilized in peptide discovery research. Purified peptides are analysed with matrix-assisted laser desorption/ionization time-of-flight/time-offlight mass spectrometry (MALDI-TOF/TOF-MS) or tandem mass spectrometry (MS/MS) systems. b, High-throughput peptidome analysis is often employed for automated sequence analysis, dereplication and/or spectral network analysis, quantitation and analysis for post-translational modifications. These analyses are usually paired with high-throughput analytical systems such as high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS). Data acquisition generates MS/MS spectra that can be analysed with de novo sequencing algorithms or used for identification of peptides with a database search. Quantitation of peptides can be achieved at the level of the mass spectrometry peak as well as with multiple reaction monitoring acquisition. Post-translational modification analysis can be performed for tryptic peptides or for endogenous peptides using a peptide fragment spectrum for database search. c, Experimental protocol and data analysis procedure for mass spectrometry imaging (MSI). After sample preparation of tissue slices (for example, mouse brain tissue), the matrix is applied and co-registered with a tissue image to precisely define sample position in the mass spectrometer. The data acquisition over the entire raster generates a summed total ion chromatogram (TIC) spectrum. To visualize m/z maps, each m/z signal can be selected to show extracted ion chromatograms. The data can be processed (normalization, smoothing, data compression) for various biocomputational analysis. Scale bars, 200 μm. Part c data processing images reprinted from ref. 133, Springer Nature Limited.