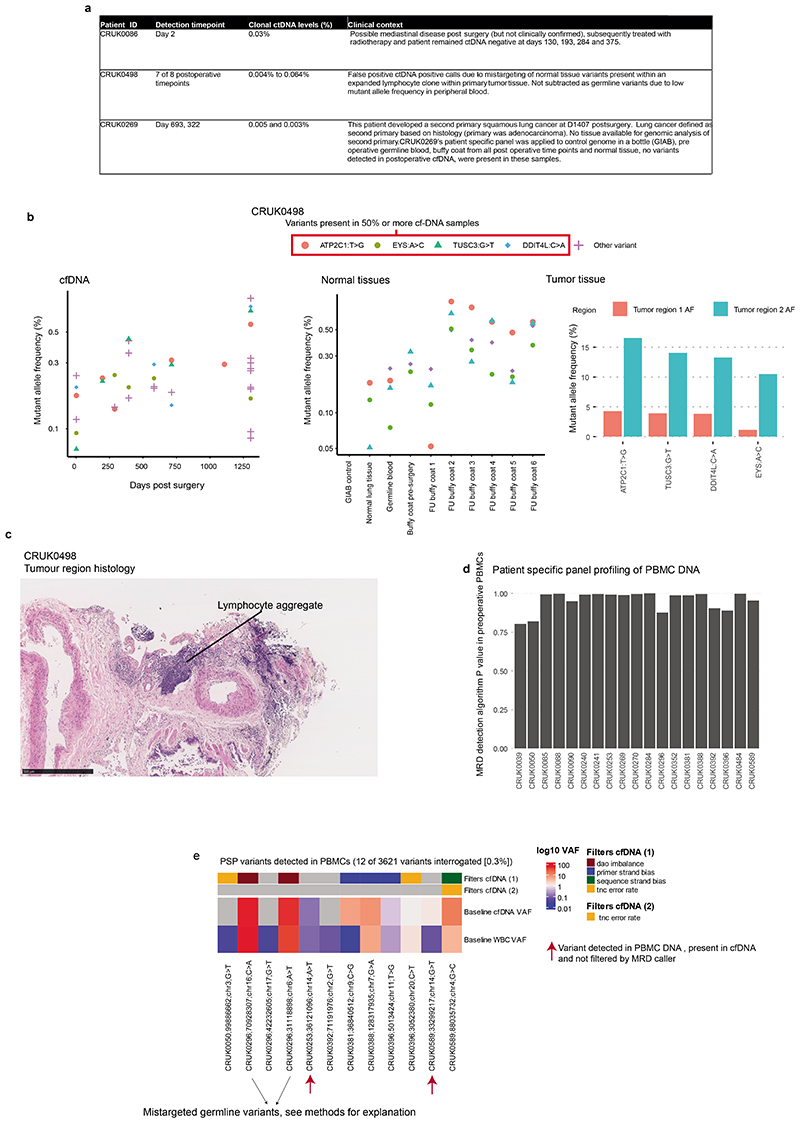
Extended Figure 5. Exploration of unexpected MRD positive results in non-relapse patients.
A. Table demonstrating details of unexpected ctDNA positive results in patients who did not suffer disease recurrence. B. CRUK0498 false positive analysis: Dot-plots represent confidently detected variants at illustrated cfDNA sampling timepoints (left panel), variants confidently detected in normal tissue, control DNA, and peripheral-blood mononuclear cell (PBMC, buffy-coat) DNA based on application of CRUK0498's patient specific panel to these respective samples (middle panel) and the mutant allele frequencies of selected variants in tumour tissue exome data (right panel). The four variants in the legend (variants in genes ATP2C1, DDIT4L, EYS, and TUSC3) represent variants confidently called at 50% or more of the timepoints across the cfDNA samples (note that confidently called means an individual variant Poisson one-sided P value of <0.01 [generated by MRD caller, see methods]). C. A haematoxylin and eosin image from patient CRUK0498's tumour where exome analysis detected the variants in genes ATP2C1, DDIT4L, EYS at high variant allele-frequencies. This image shows a dense lymphocyte aggregate in this tumour region. Scale bar below image. A single image was analysed. D. A further 19 preoperative PBMC samples were analysed from TRACERx patients; no confident panel-wide variant DNA calls were made in these patients' PBMC samples using the MRD calling algorithm. E. Variant-level analyses of the preoperative samples analysed in panel (D) highlighted that 12 of 3621 variants interrogated by the panels were detected (variant level one-sided Poisson P value <0.01). 8 of 12 detected variants were removed from the MRD caller algorithm in cell-free DNA analyses (cfDNA) due to triggering filters highlighted in the heatmap annotation. Only 2 of the 4 remaining variants carried deep alternate reads in the respective patients' preoperative cfDNA sample (red arrows). The heatmap shows the cfDNA variant allele frequency and the WBC variant allele frequency of the detected variants (grey colour represents no detection of the variant). Two mistargeted germline variants are highlighted by black arrows for patient CRUK0296, variants were targeted in error by the industry panel design pipeline but not by the TRACERx exome pipeline (methods), and were filtered from the MRD calling algorithm due to triggering the outlier filter (dao imbalance filter, dark red).