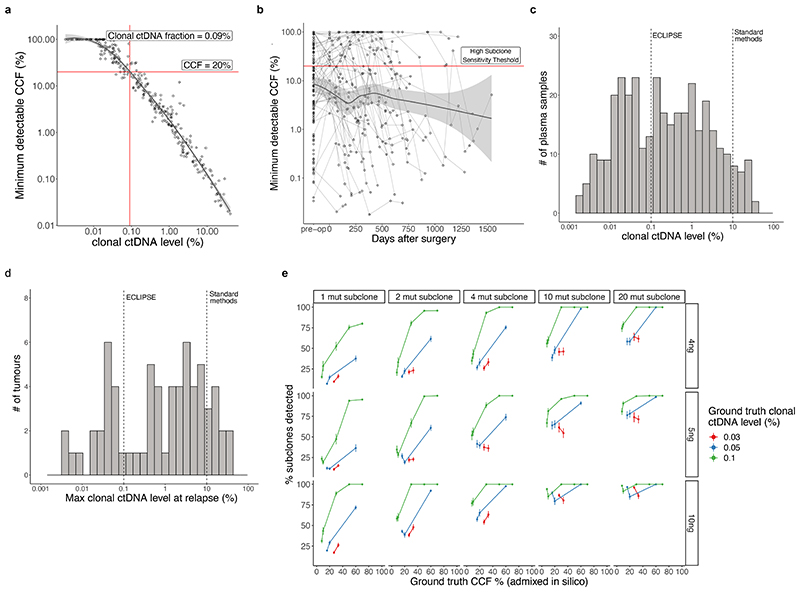
Extended Figure 8. Subclone detection sensitivity of ECLIPSE.
A. Minimally detectable CCF for each ctDNA positive sample compared to clonal ctDNA levels for each sample. All ctDNA positive samples included (N=354). Minimally detectable CCF was calculated using the minimum number of required reads for a positive (P<0.01) clone detection call (methods). B. Minimally detectable CCF over time for each patient with a horizontal line indicating the threshold for high subclone sensitivity samples (20% CCF). All ctDNA positive samples included (N=354). 61% of preoperative MRD positive samples were considered high subclone sensitivity and 66% of postoperative samples were considered of high subclone sensitivity (overall 64% of samples). C. A histogram of clonal ctDNA levels for all ctDNA positive samples (N=354) with vertical lines indicating thresholds for ECLIPSE evaluability and for traditional clonal deconvolution evaluability used for TRACERx tissue samples27 and previous clonal deconvolution approaches in ctDNA13,76. D. A histogram of maximum clonal ctDNA levels observed in post-operative samples for each patient with vertical lines indicating thresholds for ECLIPSE evaluability and for traditional clonal deconvolution evaluability (see C). This is shown for 66 patients who relapsed with ctDNA positive postoperative plasma. E. Validation of ECLIPSE detection rates across varying subclonal mutation number, clonal ctDNA level, subclone cancer cell fraction and DNA input amount into the assay. Subclones were constructed using ground truth in vitro spike-in experiments with 10-12 technical replicates for each input mass-allele fraction combination. These ground truth mutant allele fractions were then mixed in silico to construct 76,263 subclones varying across these parameters. Data from these experimentally derived subclones were then run through ECLIPSE and subclone detection rates across each of these parameters depicted.