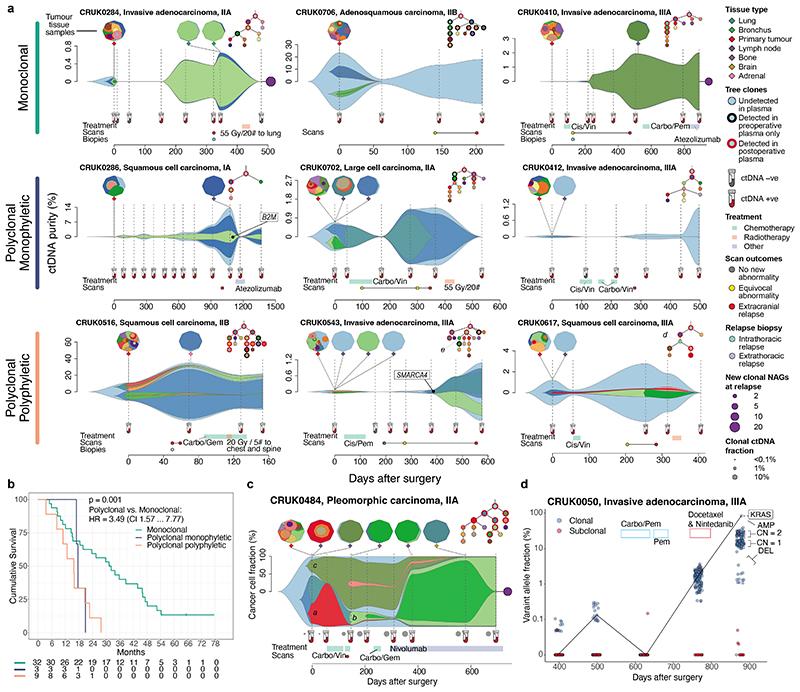
Figure 5. Longitudinal measurements of clonal evolution in plasma from surgery, through therapy and to recurrence.
A-D. ctDNA purity for each clone is calculated by multiplying the clone CCF by the ctDNA purity of the plasma sample (methods) and represents the fraction of all cells from which cfDNA was derived which harbour a given tumour clone at each timepoint. Clonal nesting is based on the phylogenetic tree for each tumour. Data from all ctDNA positive plasma samples are shown including results from ECLIPSE of samples <0.1% clonal ctDNA level. Clone maps for each tumour tissue mass are depicted above the ctDNA based clonal structure with the phylogenetic tree. Metastatic dissemination class was defined using primary tumour subclones, excluding metastatic unique clones in surgically excised lymph nodes or intrapulmonary metastases (methods). Both CRUK0617 subclone d and CRUK0543 subclone e were not detected in ctDNA but their presence was inferred by detection of its daughter subclones (Supplementary Note). A. Depictions of longitudinal tumour evolution for examples of monoclonal, polyclonal monophyletic and polyclonal polyphyletic metastatic dissemination patterns. B. A Kaplan-Meier plot depicting differences in overall survival between metastatic dissemination classes (N= 44 tumours which had at least 1 high subclone sensitivity postoperative sample). A log-rank test was used to compare survival in the two groups. C. CCFs depicted through time and therapy for CRUK0484 who experienced a polyclonal polyphyletic relapse. D. Variant allele fractions for mutations tracked in CRUK0050 at recurrence. NAG = Neoantigen, Cis = Cisplatin, Vin = Vinorelbine, Carbo = Carboplatin, Pem = Pemetrexed, Gem = gemcitabine, Gy = Gray.