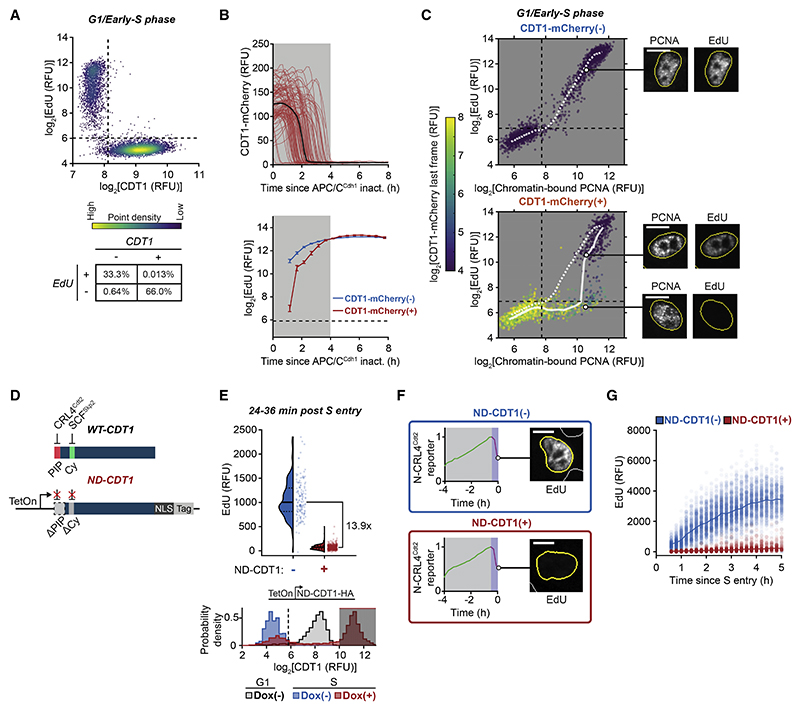
Figure 2. DNA synthesis is inhibited in the presence of CDT1.
(A) CDT1 immunofluorescence (IF) and EdU in MCF10A cells in late G1 to early S (see STAR Methods). n = 7,486 cells, representative of 2 independent experiments. Percentage of cells in bottom table. (B and C) RT-QIBC in mitogen-released MCF10A cells with APC/CCdh1 reporter and doxycycline (Dox)-inducible CDT1-mCherry. CDT1-mCherry expression in Figure S3C.
(B) Top: live-cell CDT1-mCherry (100 traces). Black curve is median trace (n = 7,059 cells). Bottom: RT-QIBC of EdU in S cells (PCNA positive) aligned to APC/CCdh1 inactivation. CDT1-mCherry(−): 18,367 cells, CDT1-mCherry(+): 6,970 cells. Error bars are mean and bootstrapped 95% confidence interval ofcells within 30 min bins (n ≥ 93 per bin).
(C) Chromatin-bound PCNA and EdU in 2N DNA cells (G1/early S), colored by live-imaged CDT1-mCherry. CDT1-mCherry(−): n = 3,000 cells, CDT1-mCherry(+): 2,000 cells. White lines are median EdU. Representative cells shown.(D–F) RT-QIBC in MCF10A cells over expressing Dox-inducible non-degradable CDT1 (ND-CDT1), induced with Dox, live-imaged for 6 h and aligned to S entry (N-CRL4Cdt2 reporter degradation). Cells born within 1 h of Dox addition were analyzed. Representative of 2 independent experiments.
(D) Diagram of ND-CDT1 construct.
(E) Top: cells 24–36 min afterSentry. ND-CDT1(−): n = 141 cells, ND-CDT1(+): n = 400 cells. Bottom: CDT1 in G1 cells (1–2 h after mitosis) (gray, n = 2,191 cells) and in S cells (0.5–1 h after S entry) with either ND-CDT1 induced (red, n = 6,389 cells) or not induced (blue, n = 783 cells). Shaded area represents cells selected for ND-CDT1(+).
(F) Representative N-CRL4Cdt2 reporter trace (magenta area represents time following S entry) and corresponding EdU image.
(G) RT-QIBC in mitogen-released MCF10Acells. Cellsweretreated with control siRNA(same experiment as Figure 3F) and induced with Dox. ND-CDT1(+) cells selected based on gating in Figure S3H. ND-CDT1(−): n = 5,500 cells, ND-CDT1(+): n = 2,000 cells. Line is median value at each time point. Representative of 3 independent experiments. Dashed lines are negative staining. Dashed and solid lines in violin plots are IQR and median. Scale bars, 10 μm.
See also Figure S3.