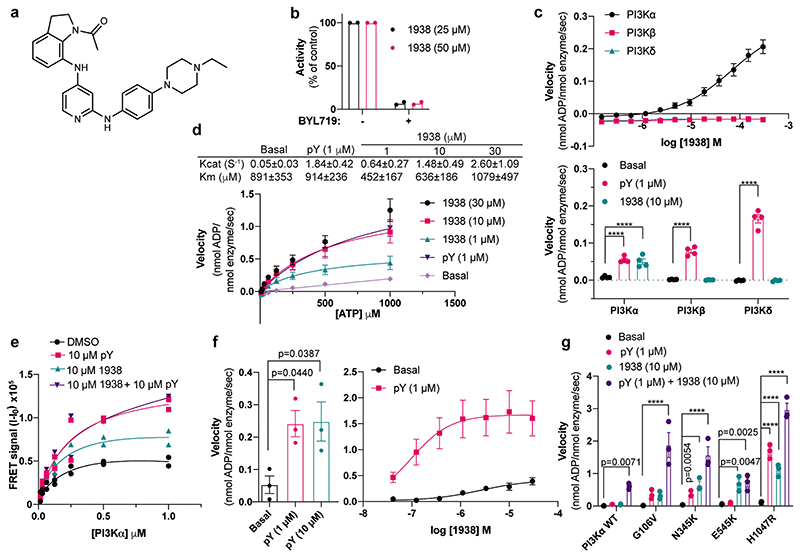
Fig. 1. Biochemical mechanism of PI3Kα activation by 1938.
a, Structure of UCL-TRO-1938 (referred to in the text as 1938). b, Effect of the PI3Kα-selective inhibitor BYL719 (500 nM) on 1938-activated PI3K. Enzyme activity in the presence of 1938 only was considered 100%. c, Selectivity of 1938 for PI3Kα over PI3Kβ and PI3Kδ. d, Enzyme kinetics (calculated using kcat function in Prism 8) upon ATP titration on PI3Kα with or without 1938 and pY. e, Membrane binding of PI3Kα shown as FRET signal (I-I0). I, fluorescence intensity at 520 nm, I0, fluorescence intensity at 520 nm in the absence of enzyme. f, Effect of 1938 on PI3Kα catalytic activity in the presence of a saturating dose of pY. g, Effect of 1938 on the catalytic activity of oncogenic mutants of PI3Kα. Data shown as n=2 independent experiments (b,e). Data shown as mean ± SEM, n=6 (c, top), n=4 (c, bottom), n=3. (d,f,g) experiments. Kinetic values in d shown as mean ± SD. Statistical analysis performed with two way ANOVA, Tukey’s multiple comparisons test (c) or Dunnett’s multiple comparisons test (g); one way ANOVA, Dunnett’s multiple comparisons test (f). ****P<0.0001.