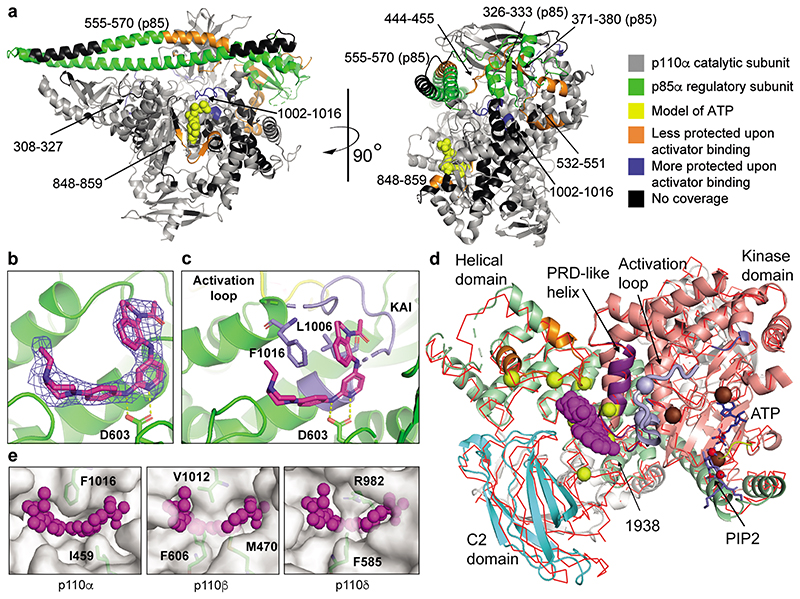
Fig. 2. Structural mechanism of PI3Kα activation by 1938.
a, Structural changes induced by 1938 as assessed by HDX-MS in full-length p110α/p85α, highlighted on the structure of p110α (gray)/niSH2-p85α (green) (pdb:4ZOP). Selection threshhold for significant peptides: a-b difference ≥2.5%, Da difference ≥0.25, p-value <0.05 (unpaired t-test). b, Sigma-weighted density map in blue (2mFo-DFc) for the 1938 ligand (magenta) in the p110α crystal structure. Yellow dashes show predicted H-bonds. c, Crystal structure of 1938 bound to p110α; 1938 (magenta), activation loop (yellow), loop 1002-1016 (kinase/activator interface, slate), predicted H-bonds (yellow dashes). d, Comparison of the 1938-bound p110α with apo-p110α. The 1938-bound structure is shown in cartoon representation, while the apo-model is shown as a superimposed red Cα trace. 1938 shown as magenta blob, PRD-like helix shown in purple. Yellow spheres mark the sites of cancer-associated mutations from the COSMIC database that are near the 1938-binding site (only mutations with >10 reports are shown). Regions showing decreased HDX-MS protection for the common helical domain mutations are colored orange. PIP2 substrate (slate) has been modelled in the active based on 4OVV. A region of the activation loop (thick worm representation, slate) has been taken from 7PG5 since it is disordered in the 1938-bound structure. Slate spheres represent residues important for PIP2 recognition (K942 and R949). Chocolate spheres represent residues essential for phosphate transfer (K776, H917 and H936). A bound ATP (blue) has been modelled based on PDB ID 1E8X. The ATP binding loop is coloured yellow. Phosphates in PIP2 and ATP are shown in red. e, Comparison of 1938-binding pocket in p110α with homologous regions in p110β and p110δ.