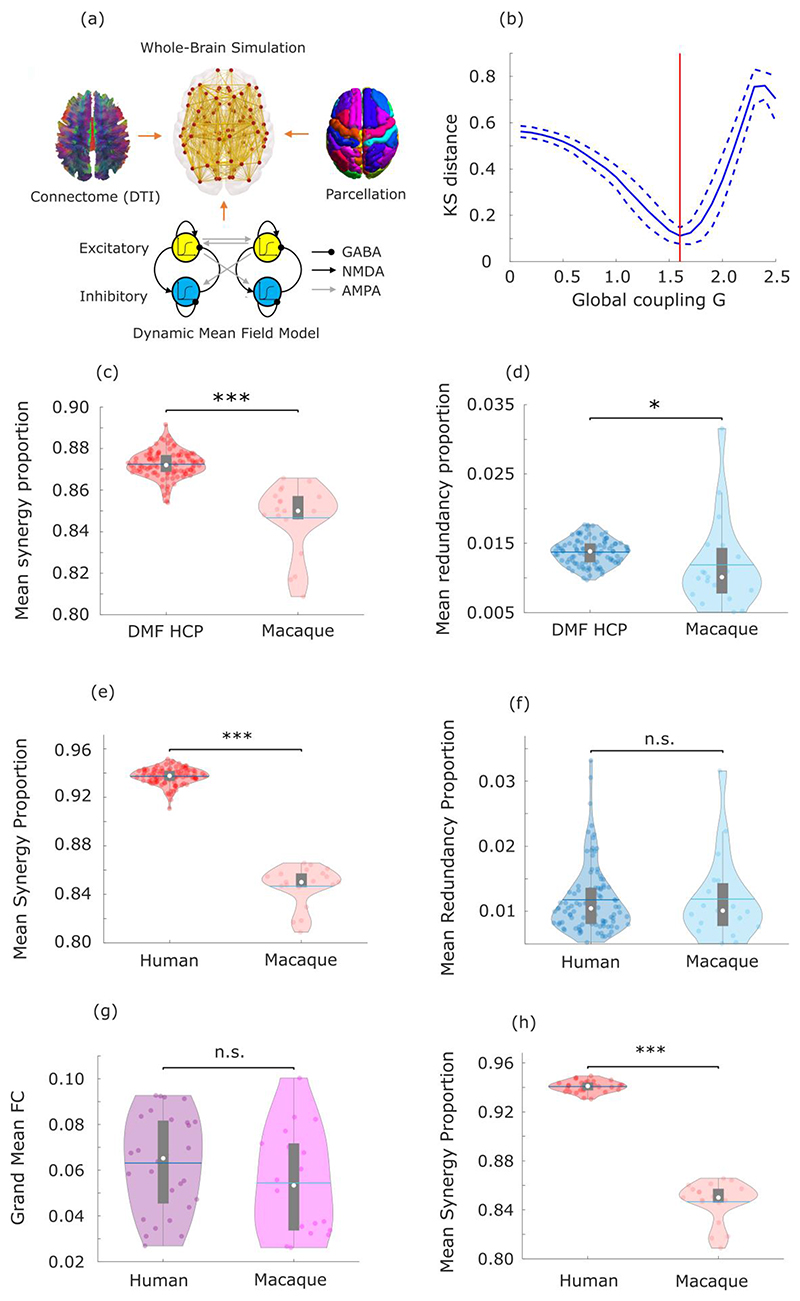
Extended Data Fig. 6. Validation analysis for human-macaque comparison of synergy and redundancy.
(a-d) Simulation of human fMRI data with same TR as the macaque data shows that human-macaque differences in synergy cannot be attributed solely to TR differences between datasets. (a) The dynamic mean field (DMF) model used to simulate human fMRI data combines macroscale information about neuroanatomy and structural connectivity (from DTI) with excitatory and inhibitory neuronal populations interconnected by AMPA, NMDA and GABA synapses, providing a neurobiologically plausible account of regional neuronal firing rate, which is turned into simulated BOLD signal by means of the Balloon-Windkessel hemodynamic model. (b) Using a TR of 0.72s (the same as the empirical HCP data), the model is fitted to the empirical HCP data by finding the value of the global coupling parameter G that minimises the Kolmogorov-Smirnov distance between the distributions of empirical and simulated functional connectivity dynamics (FCD). The KS distance is minimised for G=1.6, which is the value of G used for subsequent simulations with TR=2.6s (the same TR as the macaque data). (c) The proportion of synergistic information exchange across the brain is significantly higher in simulated human data than in empirical macaque data with the same TR=2.6s (p<0.001). (d) The proportion of redundant information exchange across the brain is also significantly higher in simulated human data than empirical macaque data (p=0.036). Statistical significance assessed with two-sample non-parametric permutation t-test (two-sided); DMF HCP data: n=100 simulations; macaque data: n=19 distinct sessions from 10 individual macaques. (e-f) The human-macaque comparison of synergy and redundancy proportion is robust to bandpass filtering both human and macaque functional MRI data between 0.008-0.09Hz. The proportion of synergistic information exchange across the brain is significantly higher in humans (p<0.001) (e) whereas the proportion of redundant information exchange across the brain is equivalent in humans and macaques (p=0.943) (f). Statistical significance assessed with two-sample non-parametric permutation t-test (two-sided). Human data: n=100 unrelated HCP subjects. Macaque data: n=19 distinct sessions from 10 individual macaques. (g-h) The proportion of total synergy is significantly higher in humans than macaques (p<0.001) (h), even when only considering humans whose total FC is in the range of values exhibited by macaques (excluding one outlier with extreme value), such that there is no significant difference in total FC between the two groups (p=0.196), shown in (g). Statistical significance assessed with two-sample non-parametric permutation t-test (two-sided). Human data: n=28 unrelated HCP subjects with FC values in the range of the macaque FC values. Macaque data: n=19 distinct sessions from 10 individual macaques (one outlier excluded in (g)). For all violin plots: each colored circle indicates one data-point; white circle: median; central line: mean; box limits, upper and lower quartiles; whiskers, 1.5x inter-quartile range; n.s., p > 0.05; * p < 0.05; *** p < 0.001.