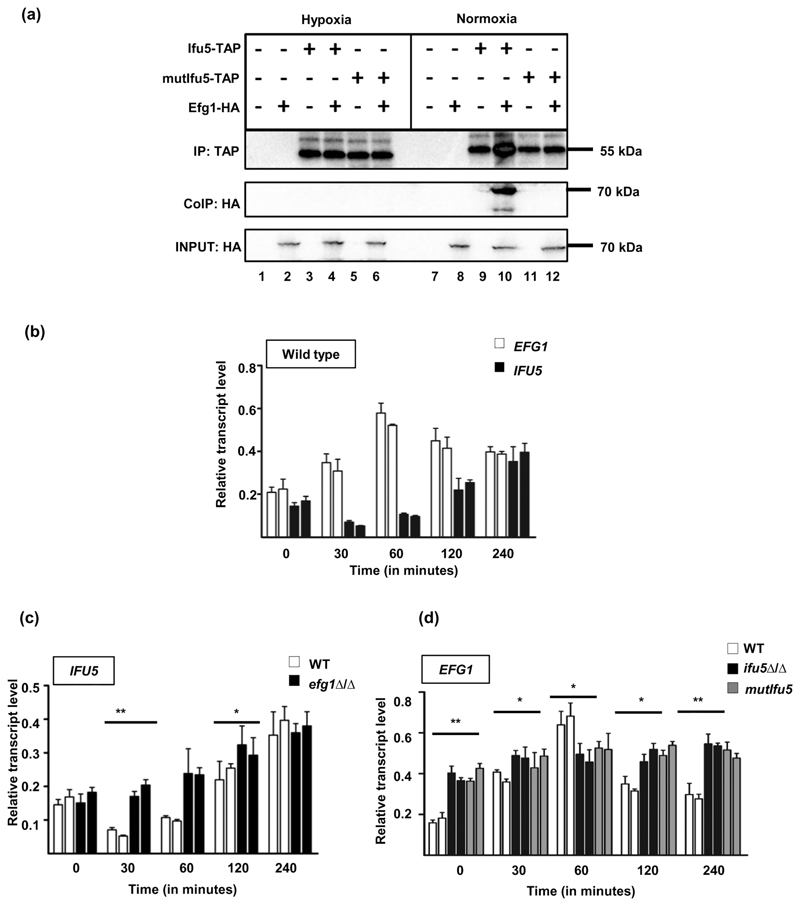
Figure 4. Role of Ifu5 in hypoxic adaptation.
(a) Strains expressing HA-Efg1 (AVL12, lanes 2 and 8), Ifu5-TAP (CLvW997, lanes 3 and 9), both HA-Efg1 and Ifu5-TAP (CLvW998, lanes 4 and 10), mutIfu5-TAP (CLvW990, lanes 5 and 11), HA-Efg1 and mutIfu5-TAP (CLvW989, lanes 6 and 12), and the control strain BWP17 (lanes 1 and 7) were grown in yeast extract peptone dextrose medium at 30°C in either hypoxic or normoxic conditions for 4 hr. The samples were immunoblotted and developed using anti-TAP (IP) or anti-HA (CoIP) antibodies (±). To verify presence of Efg1-HA, 1% of the total protein extracts was blotted and investigated with anti-HA. (b) Relative transcript levels of IFU5 and EFG1 were determined by quantitative polymerase chain reaction (qPCR). The indicated strains grown under normoxic conditions in yeast extract peptone dextrose medium for 4 hr at 30°C, were shifted under hypoxia (0.2% O2) and grown for additional 4 hr at 30°C, and 0.2% O2. RNA was isolated from cells at indicated time points. Time point 0 corresponds to 4-hr growth under normoxic conditions before shifting cells to hypoxic conditions. At each time point, two biological replicates and three technical replicates were assayed by qPCR. The ACT1 transcript was used as a calibrator to normalise IFU5 and EFG1 transcript levels. A two-tailed, unpaired t test was used to determine the statistical relevance: *p < .05, **p < .01. Relative transcript levels of (c) IFU5 transcript in wild type and efg1Δ/Δ and (d) EFG1 transcript in wild type, ifu5Δ/Δ, and mutifu5 was analysed upon shifting normoxia grown cells to hypoxic conditions. At each time point, two biological replicates and three technical replicates were assayed by qPCR. The ACT1 transcript was used as a calibrator to normalise IFU5 and EFG1 transcript levels, respectively. A two-tailed, unpaired t test was used to determine the statistical relevance:: *p < .05; **p < .01