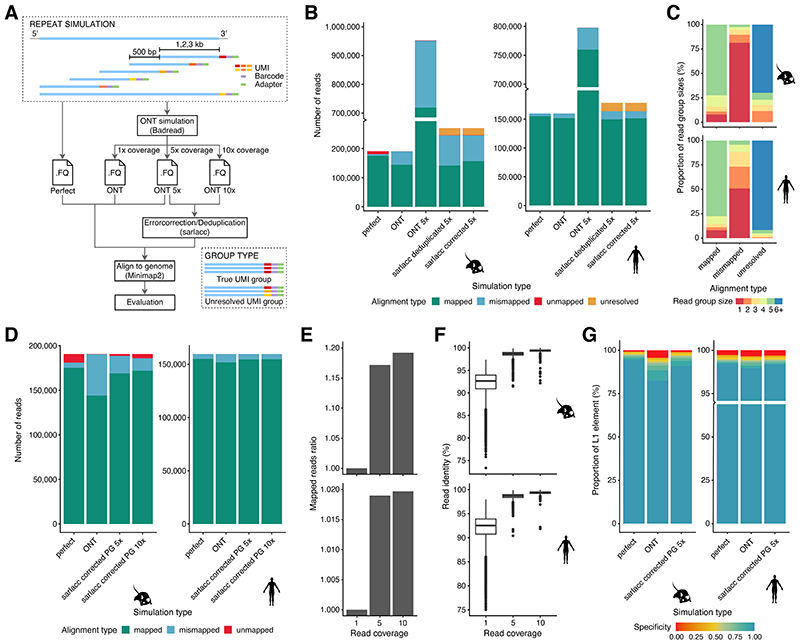
Figure 3. Simulations characterise the mapping of young L1 in mouse and human genome.
(A) schematic of simulation of reads mapping to young L1s. We simulated reads in rolling windows from the 3’ end of young L1s for the human and mouse genome. We simulated 1-3 kb read length (data for 2kb reads shown) with 500bp gap between windows and 1x, 5x or 10x read coverage. We simulated reads with perfect or ONT read identity. Reads were directly mapped to the genome or processed using sarlacc to produce deduplicated and error corrected reads (Extended Data Fig S1D). (B) bargraph of number of reads (y-axis) by simulation type (x-axis), colour-coded by alignment type. Mapped = read at correct location after mapping to the genome, mismapped = reads mapped at wrong location, unmapped = read could not be mapped to the genome, unresolved = group with multiple molecules present in one group. mouse (left), human (right). (C) bargraph of proportion of read group sizes (y-axis) by alignment type (x-axis), colour-coded by group size; mouse (top), human (bottom). (D) stacked bargraph of number of reads (y-axis) by simulation type, coloured by alignment score. (E) bargraph shows ratio of mapped reads (y-axis) by read coverage (x-axis) for mouse (top) and human (bottom). (F) boxplot of read identity (y-axis) by read coverage (x-axis) for mouse (top) and human (bottom). Mouse read coverage: 1, 5, 10 (n=140147), human read coverage: 1, 5, 10 (n=149017). The boxplots show the median, first and third quartiles as a box, and the whiskers indicate the most extreme data point within 1.5 lengths of the box. (G) Stacked bargraph showing proportion of L1s(y-axis) by simulation type (x-axis), coloured by specificity score; mouse (left) and human (right). Simulation type: perfect = perfect read identity, ONT = ONT read identity, ONT 5x = ONT read identity with 5x coverage, sarlacc corrected 5x = ONT read identity score, 5x coverage with sarlacc error correction, sarlacc corrected 10x = ONT read identity score, 10x coverage with sarlacc error correction, sarlacc deduplicated 5x = ONT read identity score, 5x coverage with sarlacc deduplication by randomly choosing 1 read. PG = perfect grouping.