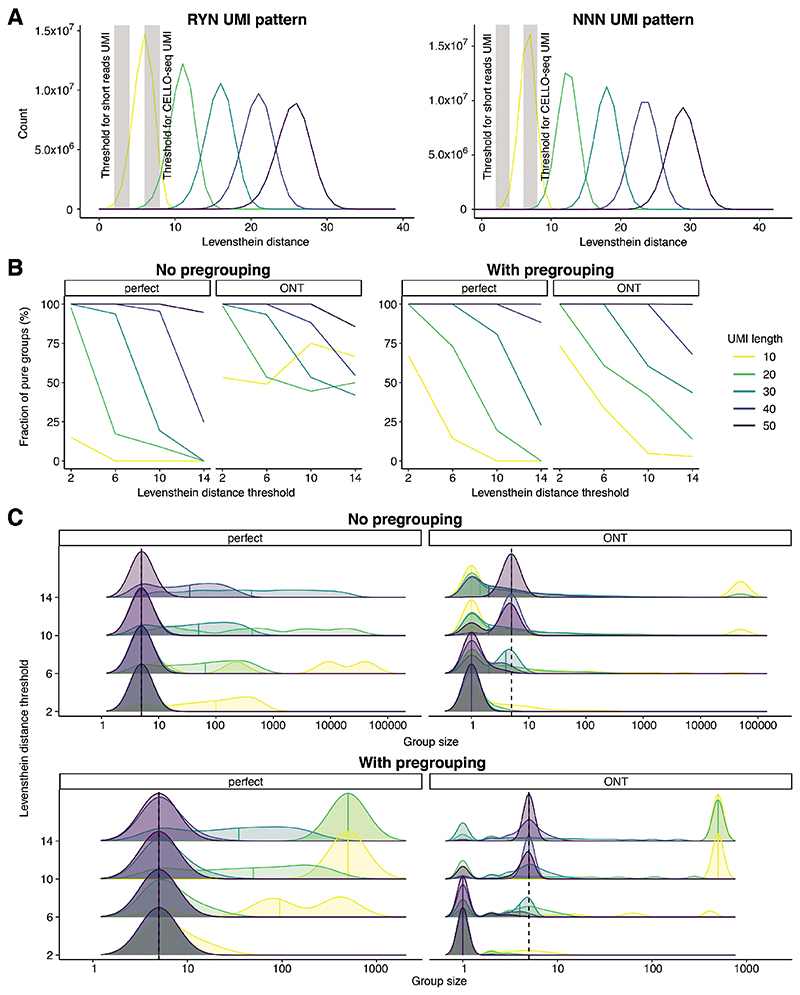
Extended Data Fig. 4. UMI simulations.
(A) Distribution of Levenshtein distance between randomly simulated UMI (x-axis) based on UMI length with RYN pattern (left) or NNN pattern (right). Light grey bar shows distance threshold for grouping of reads by UMIs used for most short read UMIs or CELLO-seq. (B) Line graph of fraction of pure groups (y-axis) by levenshtein distance (x-axis) by UMI group, either with perfect read identity or ONT read identity. On the left is the line graph of UMI simulations without any pregrouping by mapping. On the right the line graph is UMI simulation where pregrouping was performed by random assignment of true UMI sequences into groups of 100 unique UMIs. (C) distribution plot of UMI group sizes (x-axis) by levenshtein distance threshold (y-axis) based on UMI length, with perfect of ONT read identity and no pregrouping (left) or pregrouping (right).