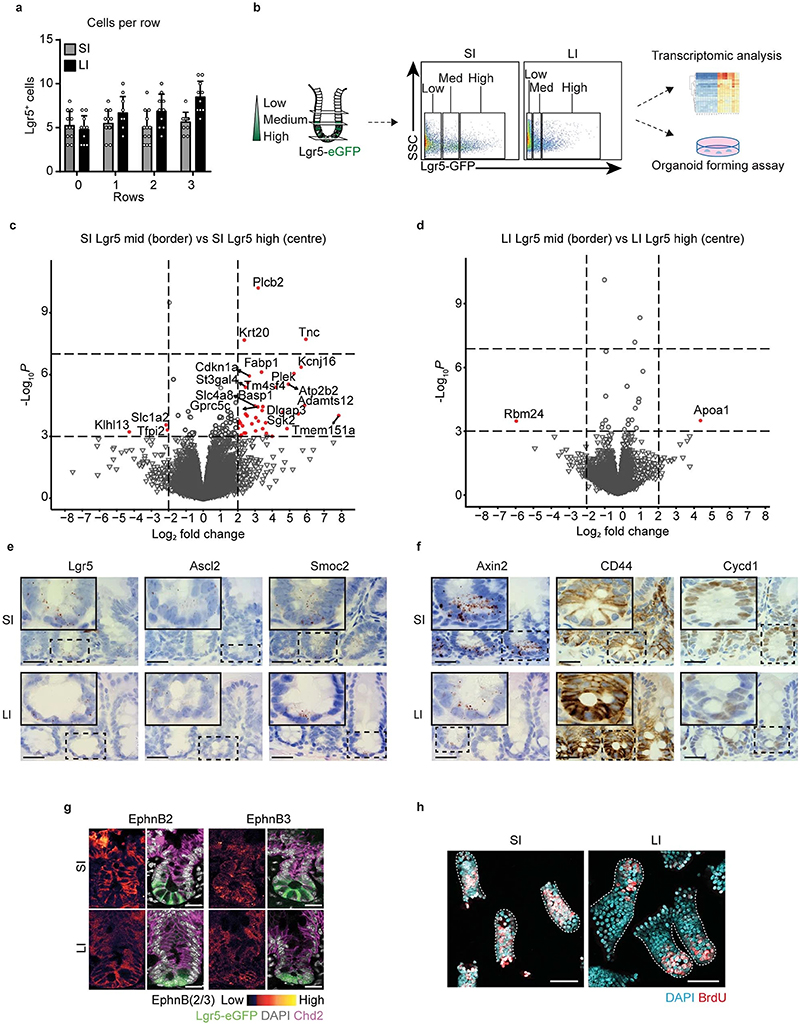
Extended Data Figure 1. Crypt characteristics in small and large intestine.
a, Quantification of the number of Lgr5+ cells per position in crypt of SI (n=12 crypts) and LI (n=12 crypts) in Lgr5eGFP-Ires-CreERT2 mice. Mean +/- SD are plotted b, Schematic representation of experimental setup for RNA-seq and organoid forming assay. c,d, Volcano plots showing log2 fold-change (x-axis) and -Log10 p-value (y-axis) of genes differentially expressed between Lgr5+ cells with medium intensity (border) and Lgr5+ cells with high intensity (centre). Genes that were significantly altered in border compared with centre Lgr5+ cells are highlighted in red (Log2 fold change >2, -Log10 p-value <0.001) in SI (c) and LI (d), n=4 mice for each condition. e, Stem cell markers (Lgr5, Ascl2 and Smoc2) in situ hybridization (ISH) in C57/B6 mouse SI (top) and LI (bottom) crypts, n=4 mice. Scale bar, 100 μm. f, Wnt targets (AXIN2, CD44, CYCD1) ISH and immunohistochemistry (IHC) in C57/B6 mouse SI (top) and LI (bottom) crypts, n=4 mice. Scale bar, 100 μm. g, Immunofluorescence (IF) staining of Ephrin B2 and Ephrin B3 in C57/B6 mouse SI (top) and LI (bottom) crypts, n=3 experiments. Scale bar, 20μm. h, Confocal images of isolated crypts (dotted outline) of SI (left), and LI (right), proliferating cells were identified by BrdU incorporation upon 2-hour pulse (red). Nuclei were labelled using DAPI (blue), n=10 experiments. Scale bar, 50μm.