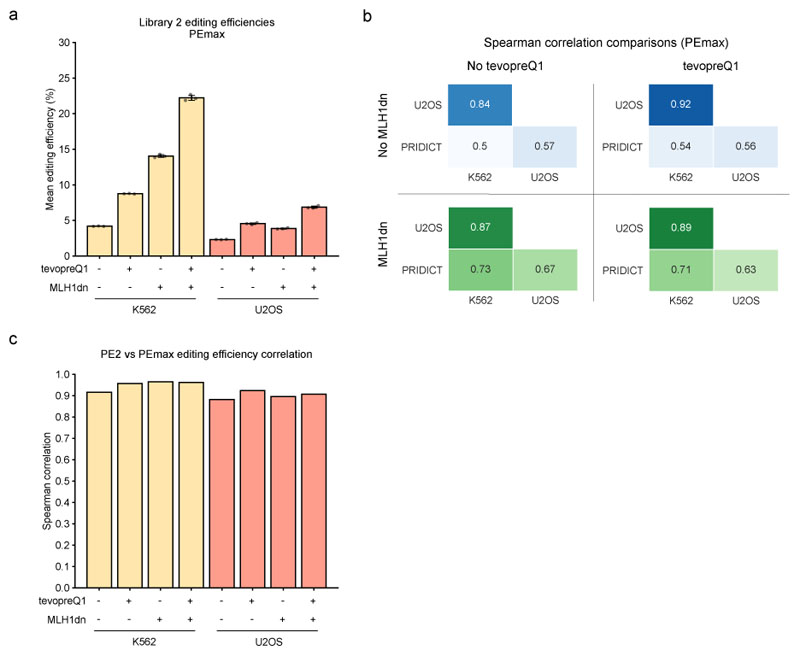
Extended Data Fig. 4. Additional library 2 evaluation with PEmax.
(a) Mean editing efficiencies of each replicate, including all pegRNAs in library 2 with different experimental conditions in U2OS and K562 cells. Error bars indicate the mean +/- SD of three biologically independent replicates. n = 3. Mean editing of library 2 for each of the 3 replicates is based on the following number of pegRNAs for each data point (bars left to right) = 916, 922, 917, 924, 879, 869, 877, 866. Note that absolute levels of editing efficiency for PEmax cannot be directly compared to PE2 in this study due to the use of different selection agents (Blasticidin for PEmax screens compared to Zeocin for PE2 screens). Previous studies showed that in identical setups, PEmax surpasses the performance of PE216. (b) Spearman correlation for PEmax editing efficiencies in library 2 between different experimental conditions (MLH1dn, tevopreQ1) and cell lines (K562, U2OS). (c) Editing efficiency rank correlations (Spearman) in library 2 between editing performed with PE2 vs. editing performed with PEmax.