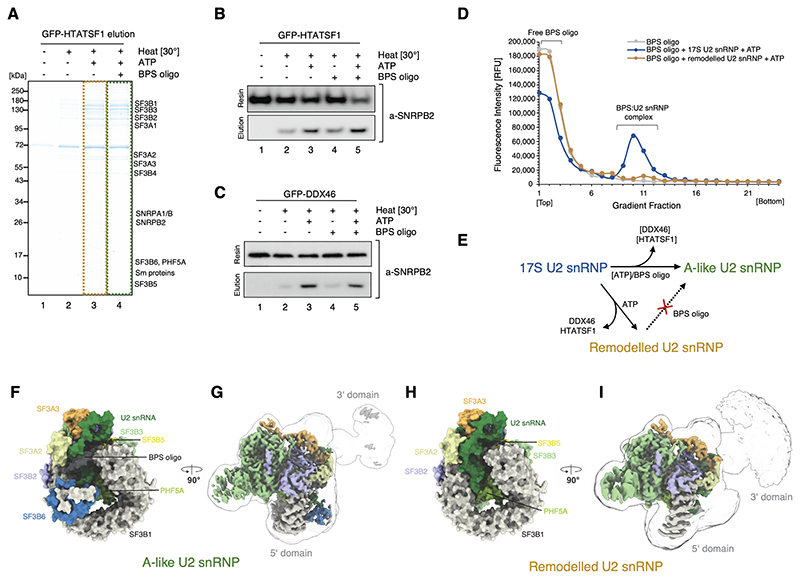
Figure 2. Sample preparation and in vitro reconstitution of the branch site recognition by the U2 snRNP.
(A) SDS-PAGE analysis of the eluates from the GFP-HTATSF1-tagged 17S U2 snRNP immobilised on the GFP nanobody resin and incubated under various conditions. (B) Western blot analysis of the reconstitution reaction performed as in (A). Elution and resin fractions were probed with antibodies against SNRPB2, a core U2 snRNP component. (C) The same as in (B), but the 17S U2 snRNP sample was immobilised using GFP tag attached to DDX46. (D) Analysis of the Cy5-labelled BPS oligonucleotide binding to the 17S U2 snRNP or remodelled U2 snRNP by glycerol gradients. RFU, Relative Fluorescence Units. (E) Schematic summarising the outcome of the in vitro remodelling and substrate binding experiments. (F) and (H) Surface representation of the 5’-domains of the A-like and Remodelled U2 snRNPs models. (G) and (I) Experimental cryo-EM maps of A-like and Remodelled U2 snRNPs showing the high-resolution 5’-domain (coloured by chain identity) embedded in a low-pass filtered (5 Å) maps, showing positioning of their 3’-domains.