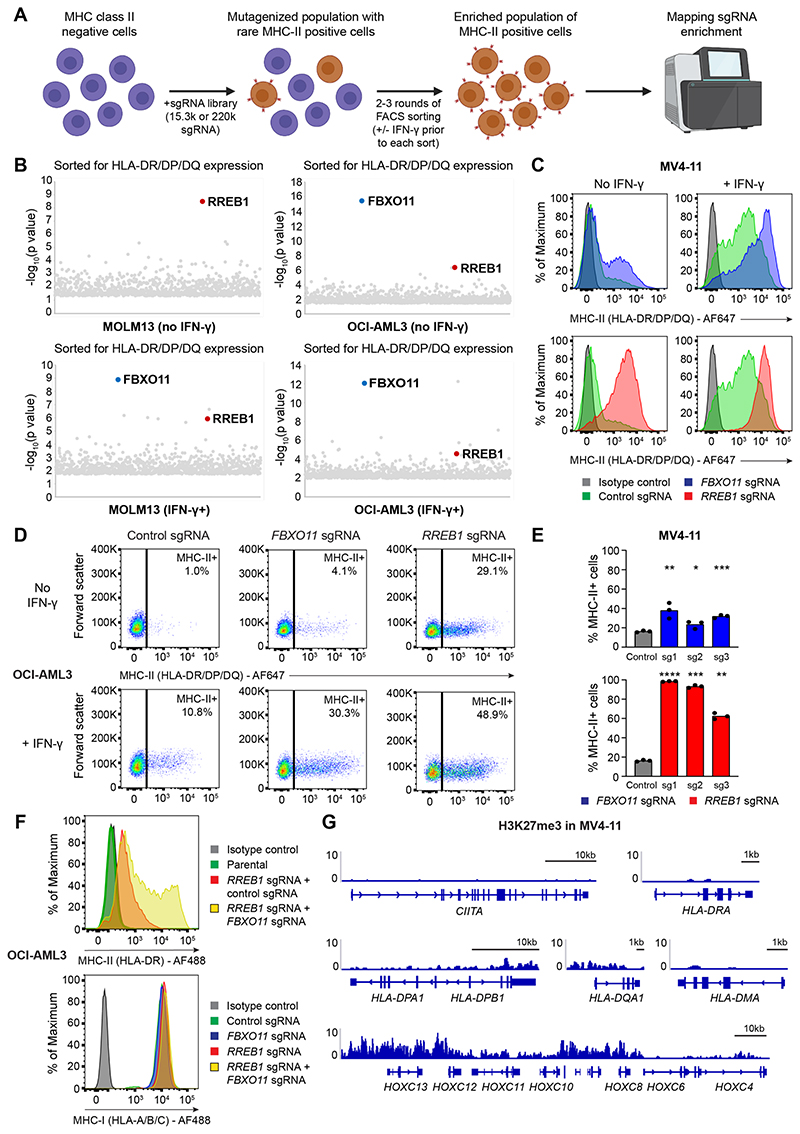
Figure 1. Genome-wide CRISPR screens identify key MHC-II regulators in AML.
(A)CRISPR screen workflow. MHC-II-negative cells (MOLM13 and OCI-AML3) were mutagenized with pooled lentiviral libraries comprising 220,000 sgRNA (whole-genome screen) or 15,300 sgRNA (targeted epigenetic screen) and MHC-II+ cells were enriched by FACS.
(B)Whole-genome CRISPR screen results. For IFN-γ+ sorts, cells were pulsed with IFN-γ 25 ng/mL for 48 hours prior to each sort. Bubble plots show the top 1,000 enriched genes identified. P values were calculated using the RSA algorithm (König et al., 2007).
(C and D) Cell surface MHC-II expression in MV4-11 Cas9 cells (C) and OCI-AML3 Cas9 cells (D) transduced with a pool of 3 sgRNAs targeting the indicated genes or a control sgRNA, in the presence or absence of IFN-γ 10 ng/mL (MV4-11) or 25 ng/mL (OCI-AML3) for 48 hours. Representative plots from 3 experiments.
(E)Cell surface MHC-II expression in MV4-11 Cas9 cells transduced with individual sgRNAs targeting the indicated genes or a control sgRNA. Bars depict the mean percentage of MHC-II+ cells from 3 independent experiments indicated by points. Statistical analysis by unpaired t test compared to control cells; p value * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001.
(F)Cell surface MHC-I and MHC-II expression in OCI-AML3 Cas9 cells transduced with sgRNAs targeting the indicated genes and/or a control sgRNA. Representative plots from 2 experiments.
(G)Chromatin immunoprecipitation (ChIP) sequencing showing H3K27me3 levels at MHC-II genes and the HoxC cluster. Data are from GEO: GSM1513828 (Göllner et al., 2017). Y axes indicate reads per million (rpm). See also Figure S1.