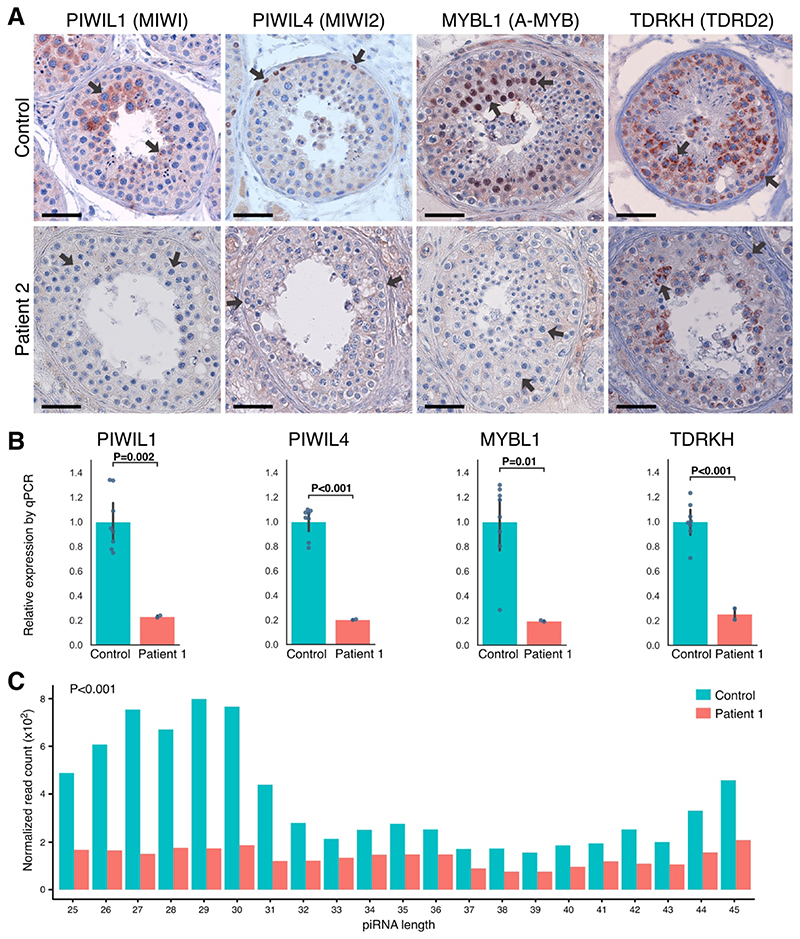
Figure 4. Expression of Key piRNA-Processing Enzymes at Protein and Transcript Levels and Small-RNA Sequencing.
Panel A shows immunohistochemical staining for P-element–induced wimpy testis–interacting RNA (piRNA)–processing proteins PIWIL1, PIWIL4, MYBL1, and TDRKH in a control biopsy sample with complete spermatogenesis and in a sample from Patient 2, who carried the biallelic missense mutation p.P84S in PNLDC1. Arrows indicate the main sites of expression. In the control, we observed expression of PIWIL1 in pachytene spermatocytes, PIWIL4 in spermatogonia, and MYBL1 in spermatocytes, which in all cases were lost or greatly diminished in the patient. TDRKH was observed in the cytoplasm of spermatogonia and spermatocytes and appeared to be less expressed in Patient 2. The expression patterns were confirmed across five controls and also in Patient 1 for PIWIL1 and PIWIL4 and in Patient 4 for PIWIL1, MYBL1, and TDRKH (Fig. S9). A testicular tissue sample from Patient 3 was unavailable for staining. The bars represent 50 μm. Panel B shows that with the use of RNA isolated from a fixed testicular tissue sample from Patient 1, the transcript levels of piRNA- processing enzymes recapitulated the pattern that was observed at the protein level. A significant reduction in transcript levels was observed for PIWIL1, PIWIL4, MYBL1, and TDRKH. The RNA from four controls with complete spermatogenesis was also obtained from fixed testicular tissue and had a similar RNA quality as in Patient 1. The analysis was performed in duplicate. A similar pattern was observed in a sample from Patient 4, but a less pronounced pattern was observed in a sample from Patient 3 (Fig. S10A and S10B). Expression of the housekeeping transcript RPS20 was similar between the patients and their matched controls (Fig. 3C). As shown in Panel C, small-RNA sequencing of RNA isolated from fixed testicular tissue sample from Patient 1 revealed a significantly lower amount of piRNAs than that in the sample from a control with complete spermatogenesis (P<0.001), as well as a major loss of piRNAs with expected lengths of 26 to 31 bases, which were evident in the control samples. Read counts of piRNAs were normalized to the spike-in RNA. Control RNA was isolated from tissue preparations that were similar to those used for the patients. Data from Patient 4 that show a similar significant reduction and from Patient 3 that show a milder effect on piRNA biogenesis are provided in Fig. S10C and S10D.