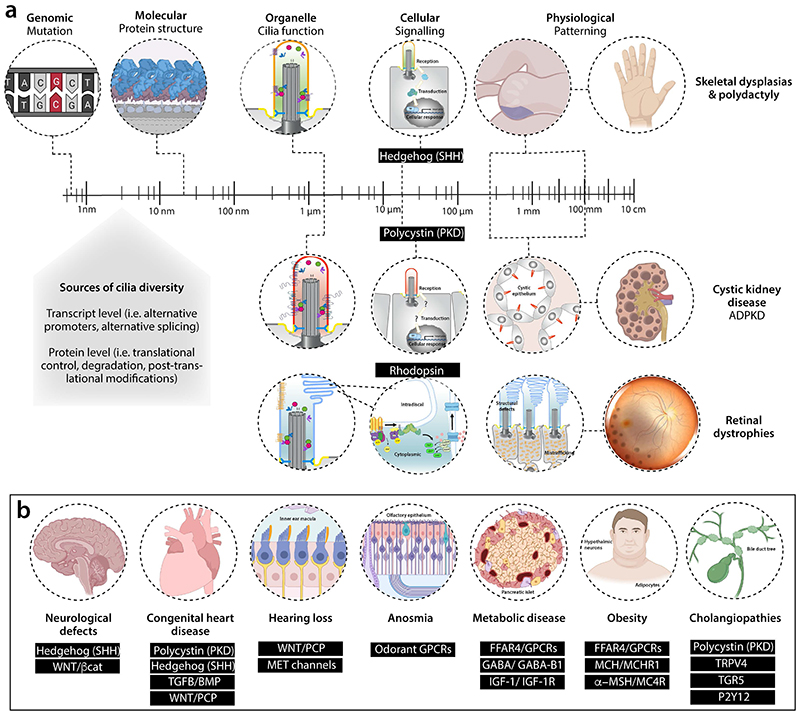
Figure 4. Challenge of ciliopathies — biology across scales.
a, Schematic of how variant identification in a patient with ciliopathy is just the start of the challenge. Genetic changes are the same across all cell types. Understanding how these variants disturb different types of cilia structurally and functionally in terms of signalling readouts is our knowledge gap across cell types and developmental times, one that we need to address to understand patient phenotypes. Possible sources of tissue-specific phenotypes are highlighted. We focus on the best characterized cilia-dependent signalling defects resulting from ciliopathy mutations in different tissue types, including skeleton, kidney epithelia and photoreceptors resulting in ciliopathic disease. For the kidney, we have focused on autosomal dominant polycystic kidney disease through PKD1/2 signalling but other renal diseases include autosomal recessive polycystic kidney disease and nephronophthisis, the latter involving additional signaling defects (see Table 1). b, Examples of the signalling pathways disrupted in different tissue types in the ciliopathies. Abbreviations: ADPKD, autosomal dominant polycystic kidney disease; FFAR4, Free Fatty Acid Receptor 4; GABA, gamma-aminobutyric acid; GPCRs, G protein coupled receptors; IGF, insulin-like growth factor; MCH, melanin-concentrating hormone; MET, mechano-electrical transduction; MSH, melanocyte-stimulating hormone; P2Y12, purinergic receptor P2Y; PKD, Polycystin; SHH, Sonic Hedgehog; TGFB/BMP, Transforming Growth Factor Beta/ Bone Morphogenetic Protein; TGR5, Takeda G protein-coupled receptor 5; TRPV4, transient receptor potential vanilloid-type 4; WNT/ βcat, canonical Wingless/Integrated Beta catenin pathway; WNT/PCP, non-canonical Wingless/Integrated Planar Cell Polarity pathway.