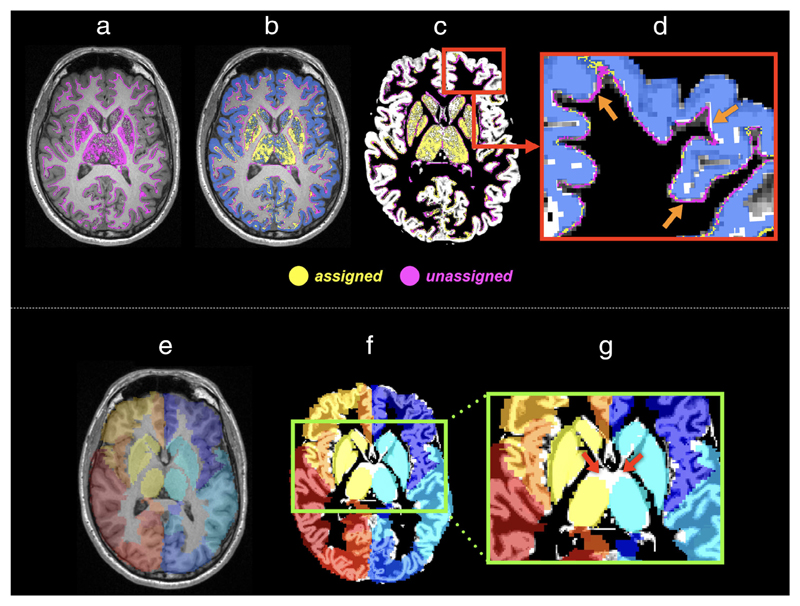
Figure 7.
The effects of the misalignment between image-intensity-based tissue segmentation and brain parcellation on the assignment of streamlines to network nodes: Upper panel: (a) With the application of anatomical constraints (e.g., Ref. 35), streamline endpoints (colored in purple) occur at the GM–WM interface or within the subcortical GM. (b) Due to factors such as discretization of structural labels, many of these endpoints (purple points) do not locate inside the, for example, FreeSurfer parcellation image (blue ribbon)48 and thus are not assigned to a label. (c) The T1 image shown in (a,b) is replaced by the GM partial volume maps derived from intensity-based tissue segmentation.80 (d) A zoom region of (c) illustrates the discrepancy (pointed by arrows) between tissue segmentation and brain parcellation, revealing that streamlines cannot be assigned purely based on the voxels where the endpoints reside.
Bottom panel: The discrepancies also present in subcortical GM, and in fact could be even crucial when parcellation images are prepared by transforming an atlas to an individual’s space (i.e., the left branch of Fig. 5). This is because the degree of misalignment between subcortical GM segmentations and brain parcellations could be increased by the registration error. As an example: (e) The AAL atlas is coregistered to individuals’ data via linear and nonlinear transformations. The background shows a T1-weighted image slice of the subject. (f) The background image is replaced by the GM partial volume map obtained from tissue segmentation. (g) The red arrows point to the considerable misalignment between two images at the thalami regions.