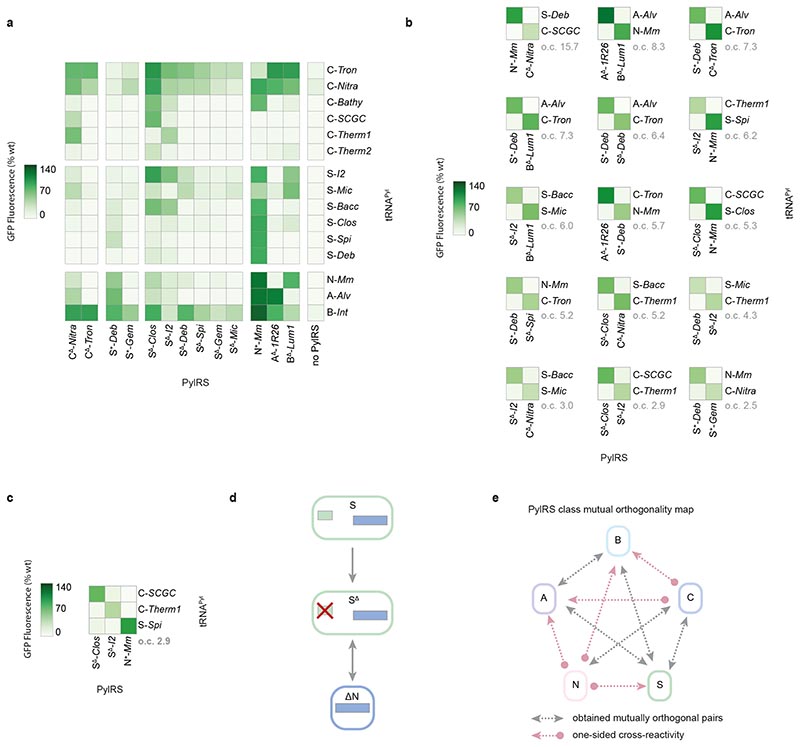
Fig. 3. Activity mapping of candidate PylRS enzymes and pyl tRNAs, and discovery of new triply orthogonal, PylRS/tRNAPyl pairs.
a. Heatmap displaying the activity of combinations of the selected pyl tRNAs and PylRS enzymes, measured by production of GFP(150AllocK)His6 from cells bearing a GFP(150TAG)His6 gene in the presence of 4 mM AllocK 1. Values are the percentage of wild-type GFP. Only PylRS enzymes and pyl tRNAs that have greater than 30% activity with at least one tRNAPyl or PylRS enzyme, respectively, are shown. Data represents the average of three biological replicates. All numerical values and bar charts including error bars showing s.d. are provided (Supplementary Table 2). b. Activity heatmaps of representative sets from each family of doubly orthogonal PylRS/tRNAPyl pairs obtained from the activity screen. Orthogonality coefficient (o.c) is shown in grey; the set with the highest o.c. in each family is displayed. Distinct members of a family share the same set of PylRS enzymes but use different pyl tRNAs. Data represents the average of three biological replicates. All numerical values and bar charts including error bars showing s.d. are provided (Supplementary Table 2). c. Activity heatmap of the set with the highest o.c. from the family of triply orthogonal PylRS/tRNAPyl pairs obtained from the activity screen. Data represents the average of three biological replicates. All numerical values and bar charts including error bars showing s.d. are provided (Supplementary Table 2). d. The generation of SΔ PylRS variants by deletion of the N-terminal domain from class S PylRS enzymes. We considered SΔ PylRS variants as engineered members of the ΔN group; their activity profiles are too diverse to be considered as a distinct class. e. The mutual interaction network between all five PylRS classes based on the activity between the characterised PylRS enzymes and pyl tRNAs. Mutually orthogonal pairs can be found using PylRS enzymes from: classes A and B; classes A and S; classes B and S; classes C and N; and classes C and S (double-headed grey arrows). Therefore, five out of ten possible mutually orthogonal combinations were discovered; the other five each showed one undesired cross reactivity (single-headed red arrows). These orthogonal interactions were identified without engineering to tailor PylRS:tRNAPyl interactions. For no two PylRS classes did all combinations of pairs possess two-sided cross-reactivity.