Summary
Biological function of macromolecules is closely tied to their cellular location, as well as to interactions with other molecules within the native environment of the cell. Therefore, to obtain detailed mechanistic insights into macromolecular functionality, one of the outstanding targets for structural biology is to produce an atomic-level understanding of the cell. One structural biology technique that has already been used to directly derive atomic models of macromolecules from cells, without any additional external information, is electron cryotomography (cryoET). In this perspective article, we discuss possible routes to chart the molecular landscape of the cell by advancing cryoET imaging as well as by embedding cryoET into correlative imaging workflows.
Introduction
Biological macromolecules exist in crowded environments inside cells, tissues, and organisms. In many cases, biological function is not only associated with the structure of individual molecules, but emerges as a result of the interactions between different molecules.1,2 An in-depth understanding of this molecular sociology not only requires high-resolution structural information about individual molecules, but also of their localization and interactions in complex cellular settings.1,3 This, in turn, necessitates the accurate mapping of macromolecules and complexes in their functional context, i.e., within the intact cellular environment.1,4
A technique ideally suited for the direct visualization of molecules in their biological environment, supporting in situ imaging within the cellular context, is electron cryotomography (cryoET).1,4–8 By combining information from two-dimensional (2D) projections of the same sample imaged at different tilt angles with respect to the electron beam, a three-dimensional (3D) volume of the sample, termed tomogram, can be computationally reconstructed. Tomograms are 3D snapshots of the specimen, typically at high nanometre resolution, allowing the spatial localization of target macromolecules within the sample. Higher-resolution structural information can be resolved via the averaging of different copies of macromolecules in the tomogram in a process called subtomogram averaging. Furthermore, cryoET supports direct correlation of the tomographic data with information obtained from a variety of associated techniques such as fluorescence microscopy and mass spectrometry (MS).
This combination of cryoET with associated techniques provides a unique opportunity to fulfill the long-standing goal to accurately and comprehensively map the macromolecular landscape of a cell. Ideally, all molecules within the cell could thus be localized and chemically identified in their native environment.
In this perspective article, we review recent developments that have brought the field closer toward this aim and discuss possible directions to be explored in the future. We identify two complementary paths that could contribute to this goal: the direct improvement of tomographic imaging and reconstruction, as well as the use of orthogonal correlative imaging techniques which allow the chemical identification of molecules within the cellular context.
Direct identification of molecules in tomograms
A tomogram is a reconstruction of a 3D cellular volume and thus, in principle, includes all the relevant spatial information about the target macromolecules. Many macromolecules, however, do not have distinct shapes or appearances that make them stand out at the typical tomographic resolution, making it difficult to distinguish these target macromolecules from the tens of thousands of other macromolecules present in the cell. Macromolecular localization in tomograms is hence directly linked to tomogram quality and resolution, as better tomograms allow distinguishing between more molecular species. Therefore, increasing tomogram quality and resolution is an important future area of development for the field.
There are several intricately interrelated factors that limit tomogram resolution. The most important and fundamental limitation is radiation sensitivity of biological specimens.9 Radiation sensitivity leads to restrictions in the usable dose and thus in achievable signal-to-noise ratio, which reduces contrast and complicates the alignment of the tilt series images. An accurate alignment is, however, essential for high-resolution tomogram reconstruction. This limitation is less severe in the imaging of inorganic specimens, where significantly higher doses, and thus resolutions, are possible.10 Other resolution-limiting factors are the increased sample thickness of specimens prepared to retain a biologically intact sample state, which correlates with an increasing number of multiply scattered electrons and thus further affects the signal-to-noise ratio, and unavoidable reconstruction artifacts due to missing tilt angles, i.e., the “missing wedge”.
Given that the 3D information contained in a tomogram is retrieved from the information stored in each of the images acquired at different tilt angles, an artefact-free, high-resolution tomographic reconstruction in principle requires imaging at the full angular range, sampled as finely as possible to completely map out the corresponding 3D Fourier space.11 As, in practice, both the number of obtainable tilt series images as well as the accessible angular range are limited by radiation damage effects and sample geometry,1,2 these factors, especially dose, number of projections, and sample thickness, have to be carefully balanced to obtain high-resolution tomograms.
Given the diversity of interconnected factors currently limiting tomogram quality and resolution, there are several levels on which technical advances could lead to significantly improved tomograms, ranging from improvements in sample preparation and hardware to innovations in data processing and optimisations in the data acquisition process.
Improvements in sample preparation and microscope hardware
The interaction of the electron beam with the sample can lead to significant beam-induced motion of the target molecules, which is often associated with radiation-related distortions of the ice layer that is not perfectly planar.12,13 Specimen movement leads to blurring and inaccurate tilt series alignment and can even induce changes in the specimen during tilt series acquisition, resulting in an irrecoverable loss of resolution.14 Thus, tomogram quality and resolution could be significantly improved by the reduction of specimen movement during data collection. On the specimen preparation side, the use of ultrastable gold supports that limit radiation-induced specimen movement13,15,16 is a proven method to improve tomogram quality.14 Further development in specimen supports is likely to improve tomographic reconstruction and is the subject of research in several laboratories.
In addition, sample thickness is a major limiting factor in cryoET, because increased sample thickness leads to an increase in inelastic scattering events during imaging, resulting in lower signal-to-noise ratios in the collected tilt series images. Preparation of thinned specimens, especially when working with cellular samples, alleviates this problem. Thin samples can be obtained using a variety of techniques that range from targeting thin edges of cells (such as filopodia), to the use of genetic mutants producing smaller cells (sometimes called mini-cells), and to the sectioning of frozen specimens using a diamond knife followed by subsequent imaging (CEMOVIS).17–21 In recent years, the adaptation of focused ion beam (FIB)-milling for biological specimens22,23 has substantially advanced cryoET imaging. FIB-milling employs an ion beam to ablate material from the specimen above and below the target area, resulting in thin lamellae with a thickness of 100–250 nm containing the sample volume of interest. While FIB-milling can lead to structural damage to the molecules near the surface of the lamellae and thereby might restrict the achievable resolution, in many cases, sample thickness is the more dominant factor causing deterioration of signal-to-noise ratio.24 FIB-milling is thus a controlled technique for producing thin samples suitable for cryoET with negligible damage to the interior of the specimen, which is applicable to a wide range of cellular systems.22–34 Recent advances in automation of lamellae milling23 have dramatically increased the throughput of this promising method. Further automation of this important technique will result in higher quality cellular tomograms for macromolecular mapping.
Complementing the developments in sample preparation, additional advances on the microscope hardware side that have the potential of improving tomogram data include imaging at helium temperature, which has been shown to reduce radiation damage to the specimen,35 and chromatic aberration correction36,37 which can enhance the imaging of thicker samples in which inelastic scattering processes are significant. The chromatic aberration of the objective lens results in the inability to focus electrons of different energies (i.e., different wavelengths) onto the same focal plane. Thus, inelastically scattered electrons, which lose energy during the interaction with the specimen, are not correctly focused. These electrons either increase noise in the micrographs or are discarded by the use of an energy filter, not providing any useful information about the specimen. Chromatic aberration (Cc) correction would correct for this and hence allow inelastically scattered electrons to contribute to the image and thereby improve the signal-to-noise ratio of tomograms, which are inherently restricted by the dose limitation characteristic of biological specimens.36,37
Developments in detector technology and sensitivity can further improve the quality of tomographic imaging. Direct electron detectors allow for faster data acquisition by acquiring rapid image sequences instead of single images, thereby reducing specimen movement within tilt images, and supply images with substantial information transfer even at the sampling limit, i.e., the Nyquist frequency.2,38,39 In addition, an increased imaging speed boosts throughput, especially when combined with faster tilt schemes. Some examples of such tilt schemes include continuous tilting of the goniometer combined with a rapid acquisition of tilt frames,40 fast-incremental tilt schemes41 or parallelized acquisition of multiple areas of the specimen using beam image shifts.42 As the use of direct electron detectors has been a major factor in enabling electron cryomicroscopy-based structural biology up to atomic resolution, further improvements of detector performance, for example by an increased detective quantum efficiency and a higher accuracy in the subpixel localization in electron counting, could be an important contribution to increasing tomogram quality and resolution.43,44
Advances in data acquisition and tilt series data processing
Another route toward tomogram enhancement is the optimization of data acquisition and data processing schemes. Data collection and processing are tightly connected, and therefore must be considered together for any tomographic experiment. Improvements in these can boost resolution and reduce artifacts induced by physical limitations of the imaging process discussed previously.
The tilt scheme used for acquiring the tomographic data has a significant effect on the quality of the tomogram, because it determines in which angular region the highest quality data can be obtained, as the number of obtainable projections is strictly dose-limited. Indeed, only few tilt images contain high-resolution information required for high-resolution structural biology, since this information quickly gets lost due to radiation damage. Thus, the implementation of a dose-symmetric tilt scheme,45 which begins acquisition on the untilted—and hence thinnest—sample, and gradually employs higher and higher tilts while alternating between positive and negative tilt angles, has been shown to maximize the obtainable resolution, as estimated by subtomogram averaging.46 Such a scheme is now standardly used in many cryoET applications. Advances in achievable resolution offered by the dose-symmetric tilt scheme could be further improved by combining it with other modified tilt schemes which have shown the potential for an overall improvement of tomographic reconstruction, such as equally sloped tomography (EST).11 EST is a tilt scheme that aims at the optimization of the mapping between the set of tomographic projections, which are described in polar coordinates, and the reconstructed object described in Cartesian coordinates, in order to reduce artifacts and improve the quality of the reconstruction. This is realized by using pseudopolar fast Fourier transforms, which provide an exact mapping from a Cartesian grid to a pseudopolar grid featuring equally sloped lines rather than the equally angled lines of a polar grid. These mathematical considerations can be implemented into the tomographic imaging process by combining pseudopolar Fourier transforms with a tilt scheme that yields equally sloped tomographic projections in pseudopolar coordinates. Hence, instead of acquiring projections at equal increments of the tilt angle (θ), EST uses a set of tilt angles determined by equal steps of the slope of the tilt angle (tan θ). This scheme, combined with an iterative oversampling algorithm, has been shown to improve tomogram resolution as well as dose-efficiency in several applications, achieving better than 4 Å-resolution for electron tomography of inorganic specimens.10,47–51 Advances on the data acquisition level can thus substantially improve tomographic imaging, especially if they have the potential for increasing dose efficiency. Another potential method to increase tomogram quality is the use of dual-axis tomography, which collects tilt series in two orthogonal tilt axes. Dual-axis tomography has been shown to reduce wedge-related artifacts and thus increase the quality of tomographic re-constructions.52–54 For radiation-sensitive samples, however, employing a dual-axis scheme has to be carefully balanced with dose considerations, and applying this method is cumbersome, involving grid rotations in the microscope, leading to limited reported applications in the last few years.
Tomogram quality can additionally be improved by using computational solutions that correct for remaining artifacts. Alignment accuracy could for example be increased by integrating multi-particle refinement schemes in subtomogram averaging,55 which improve tilt series alignment by correcting for beam-induced motion. Other studies have reported improvements by employing center-of-mass-based alignment schemes.10 In addition, mathematical concepts such as compressed sensing,56,57 which exploits the sparsity of a signal to reduce the number of necessary measurements to recover it, may be employed to reduce artifacts in cryoET reconstructions,58–63 and can allow improvements in signal-to-noise ratio and resolution of the resulting tomograms without the need for additional data collection.
Improvements in template matching techniques and tomogram processing
In order to draw conclusions about a molecule of interest, it must first be localized within the densely packed cellular environment observed in tomograms. A technique that has been successfully employed to aid with particle identification within tomograms is template matching.64,65 Template matching computationally identifies target molecules within a tomogram by comparing voxels within the tomographic volume with a template. The choice of template depends on the a priori knowledge available about the target molecule. A typical example would be lowpass-filtered features of a known atomic structure of the molecule of interest.64,65 Additional steps, such as segmentation or denoising, can further improve structure identification and reduce the number of false positives detected by the template matching process.6,66,67 This automated way of localizing targets within a tomogram provides a visualization of the spatial distribution of the macromolecules of interest, hence subsequently applying the method with a range of templates could delineate the 3D arrangement of the different cellular components. Template matching is thus a valuable asset in assembling a complete map of the cellular landscape.
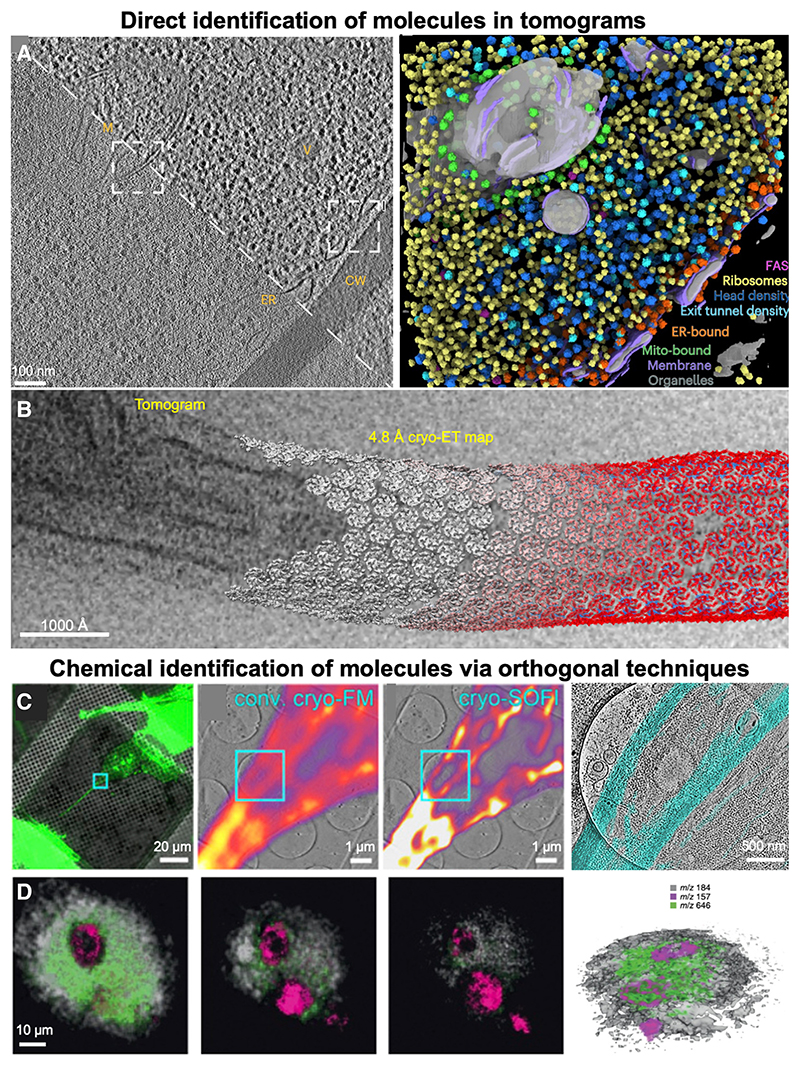
Recent developments in template matching include de novo template generation and template-free pattern detection, i.e., the localization of target molecules from the tomographic data alone, without any external references or templates.6,68,69 Other developments are the inclusion of deep learning- and neural network-based approaches to pattern mining and template generation, which have significantly advanced the accuracy and the speed with which patterns can be detected in tomograms,68–73 especially when combined with the computational prediction of potential protein-protein interactions.74–76 For example, macromolecular location prediction within a tomogram using the convolutional neural network-based software DeePiCt led to the identification of macromolecular complexes of varying shapes and sizes (Figure 1A).72
Figure 1. Techniques for localizing macromolecules within the cellular environment.
(A) Mining molecular patterns in tomograms of cellular volumes using neural network-based picking. Left: 2D slice of a tomogram of Schizosaccharomyces pombe, raw data (below dashed line) and contrast-enhanced by an amplitude equalization filter (above dashed line). Discernible organelles are labeled: mitochondrion (M), vesicle (V), endoplasmic reticulum (ER) and the cell wall (CW). Right: Macromolecular localization prediction for the cellular volume by the convolutional neural network-based software DeePiCt. Figure adapted from ref.72
(B) Employing high-resolution maps obtained by subtomogram averaging for localizing macromolecular structures in cellular tomograms. Slice through the tomogram of a Caulobacter crescentus cell overlaid with copies of the 4.8 Å-resolution structure of the S-layer, composed of repeating copies of the RsaA protein, from subtomogram averaging in the refined locations. Figure adapted from ref.83
(C) Super-resolution cryoCLEM of mammalian cells. Left to right: Overlay of conventional cryo fluorescence microscopy (cryo-FM, green) and reflected light (gray) overview images of mammalian cells with Dendra2-Lifeact-labeled actin; overlays of tomogram slices of the area indicated in the left image with conventional cryo-FM (center) and with cryogenic super-resolution optical fluctuation imaging (cryo-SOFI); overlay of corresponding tomogram sections with actin-containing volumes (cyan). Figure adapted from ref.93
(D) 3DFIB-SIMS imaging of a rat alveolar macrophage cell incubated with the drug amiodarone. Left to right: Sequence of time-of-flight SIMS images of the sample as the cell is milled by a Bi3+ ion beam showing the distribution of phosphocholine marker (gray), nuclear marker (magenta), and amiodarone (green) in different layers of the specimen, and 3D rendering of the corresponding volume. Figure adapted with permission from ref.114
The ability to locate individual macromolecules opens up another important possibility of tomogram resolution improvement via subtomogram averaging structure determination, which we discuss briefly for completeness. In recent years, the resolution of macromolecular structures obtained from tomographic data has been pushed beyond previous limits by employing subtomogram averaging.14,77–83 Subtomogram averaging requires the extraction of subvolumes containing copies of the target molecule from a tomogram and their subsequent 3D alignment and averaging analogously to single-particle analysis for cryoEM.1,79 The averaging process enhances contrast, and as the orientations of the individual molecular copies differ with respect to the missing wedge, anisotropy in the reconstruction is reduced. Subtomogram averaging can thus significantly improve the resolution of the resulting molecular structure determined from target molecules in the cellular environment and also reveal the orientation and interactions of the molecules, which are important for building the molecular picture of the cell, including macromolecules in the cytoplasm or molecules forming ordered superstructures in cells (Figure 1B). Subtomogram averaging has been successfully applied to a range of macromolecules including ribosomes,84 virus particles,85,86 bacterial or archaeal surface layers,87,88 proteasome complexes6 and membrane proteins.89 As subtomogram averaging relies on target identification for subvolume extraction, subtomogram averaging and template matching are highly synergistic methods that can be combined to yield efficient structure determination workflows for macromolecules in the cellular context.
In summary, cellular tomographic data not only contains structural information about classes of molecules, it also contains information regarding the localization of said molecules, as well as their orientation relative to one another. As the cellular environment is densely packed with a range of different molecules, identifying molecules of interest is crucial both for subtomogram averaging and for studying interactions of molecules within their native environment.
Chemical identification of molecules in cells using orthogonal techniques
While the direct improvement of tomogram quality and resolution will be crucial for the creation of a full molecular atlas of the cell, correlation of cryoET data with molecular localization data from different imaging approaches, especially methods exhibiting chemical sensitivity, has the potential of significantly contributing to this goal.
A correlative imaging technique that has been successfully applied to a range of cellular processes is cryo-correlative light and electron microscopy (cryoCLEM).21,90 CryoCLEM combines analysis by cryoET with a fluorescent light microscopy imaging step performed at cryogenic temperatures. Cryo-light microscopy provides a map of all fluorescently tagged molecules, which can then directly be correlated with cryoET data91 (Figure 1C). This process of correlation is often aided by the use of fiducial markers visible in both light and electron microscopy.91 As cryoET itself is not a chemically sensitive technique, prior identification of the target molecules via fluorescent tagging permits molecular localization in tomograms.21 Molecular identification and localization in cryoCLEM can further be improved by utilizing advances in light microscopy resolution such as super-resolution techniques92,93 (Figure 1C) or single-molecule localization microscopy.94–96 Given that not all molecules of interest can be tagged fluorescently, alternative tagging strategies could be used to facilitate molecular localization within the cell. As metals appear at higher contrast in electron micrographs, metal-containing molecules such as gold-antibody conjugates,97 metallothionein,98 and ferritin99 could be used as tags in electron microscopy imaging. Within the framework of electron microscopy, electron energy loss spectroscopy (EELS) and cryo-scanning transmission electron microscopy (cryo-STEM) can also provide insights into the chemical identity of the imaged objects.100–103 In the case of EELS, the unique energy loss spectrum of each element is exploited for chemical identification, while in cryo-STEM, the scattering angle of the incident electrons is a function of the atomic number of the chemical element, which can be utilized for identification.
To gain enhanced chemical sensitivity without the need for tagging, cryoET can be combined with MS-based techniques. When studying macromolecules and their interactions in the cellular context, one option is the correlation of high-resolution spatial data obtained from cryoET with cross-linking MS.104,105 As cross-linking MS provides information about expected interactions via the formation of additional bonds between residues in close proximity, this data can help with the interpretation of unexplained densities or provide guidance when searching for the relevant macromolecules and their interaction partners in the tomogram volume.105 The prediction of protein-protein interactions can also significantly be aided by artificial intelligence-based methods,74–76 which can be integrated into correlative workflows including template matching and cross-linking MS. Specific interactions, especially those involving small molecules or ligands, can also be explored using native MS,106–108 which can further guide interpretation of tomograms. In addition, native MS-based techniques could be integrated into the sample preparation process for electron microscopy imaging of purified biological samples,109,110 pointing toward possible fruitful future developments in combined MS and electron microscopy workflows.
Another MS-based technique with potential for correlative imaging with electron microscopy is focused ion beam secondary ion mass spectrometry (FIB-SIMS). In FIB-SIMS, the secondary ions created by focused ion beam milling of the sample are analyzed using a mass spectrometer,111–113 thus the technique directly combines spatial information gathered by FIB/SEM with chemical information from MS. This approach hence has the advantage of allowing nearly simultaneous spatial and chemical imaging. As material is continuously ablated during the FIB-SIMS process, the method also has access to the 3D sample volume. 3D imaging by FIB-SIMS has been successfully applied to identify small molecules in tissue specimens (Figure 1D). While FIB-SIMS is an established technique for elemental analysis in materials science,111,113 the FIB-SIMS analysis of biological samples is more complex, as larger molecules need to be identified via their fragmentation pattern. However, developments in recent years such as the use of gas cluster ion beam sources and advanced mass spectrometers have facilitated an increase in the obtainable spatial and mass resolution as well as the identification of larger molecules in biological samples.112,114–116 Combined with lamella preparation and cryoET, FIB-SIMS is a promising technique, which could provide information about both the chemical identity and the spatial localization of molecules within their native cellular environment.
Conclusion
The feasibility of structural biology in situ has been demonstrated by multiple developments in cryoET discussed in this article, culminating in the high-resolution structure determination of a growing number of macromolecules directly from the cellular environment. While some parts of the cell, such as the nucleus, as well as molecular complexes with low abundance, will be very challenging to map, the continuous improvement of the methods employed for this purpose could, in the future, contribute to the generation of a full atlas of the cellular landscape. The two approaches outlined in this article, namely the overall improvement of tomogram resolution and the chemical identification and localization of molecules in the cell using correlative techniques, are both promising strategies for mapping the molecular landscape of the cell. The two approaches are harmonious and the best results will likely be obtained if they are employed in parallel. In summary, the future of structural biology is bright; as techniques for whole cell investigations mature, the possibility of charting the molecular landscape of the cell will soon become a reality.
Acknowledgments
This work was supported by the Medical Research Council, as part of United Kingdom Research and Innovation (also known as UK Research and Innovation) [Program MC_UP_1201/31]. For the purpose of open access, the MRC Laboratory of Molecular Biology has applied a CC BY public copyright license to any Author Accepted Manuscript version arising. T.A.M.B. would like to thank the Wellcome Trust (grant 223788/Z/21/Z), the Human Frontier Science Program (grant RGY0074/2021), the European Molecular Biology Organization, the Leverhulme Trust, and the Lister Institute for Preventative Medicine for support. The authors would like to thank Lori Passmore and Jan Böhning for critically reading this manuscript.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Beck M, Baumeister W. Cryo-Electron Tomography: Can it Reveal the Molecular Sociology of Cells in Atomic Detail? Trends Cell Biol. 2016;26:825–837. doi: 10.1016/j.tcb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Asano S, Engel BD, Baumeister W. In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. J Mol Biol. 2016;428:332–343. doi: 10.1016/j.jmb.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 4.Baumeister W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol. 2002;12:679–684. doi: 10.1016/S0959-440X(02)00378-0. [DOI] [PubMed] [Google Scholar]
- 5.Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular Architecture in Eukaryotic Cells Visualized by Cryoelectron Tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 6.Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R, Baumeister W. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- 7.Dobro MJ, Oikonomou CM, Piper A, Cohen J, Guo K, Jensen T, Tadayon J, Donermeyer J, Park Y, Solis BA, et al. Uncharacterized Bacterial Structures Revealed by Electron Cryotomography. J Bacteriol. 2017;199:e00100–e00117. doi: 10.1128/JB.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhning J, Bharat TAM. Towards high-throughput in situ 2021 structural biology using electron cryotomography. Prog Biophys Mol Biol. 2021;160:97–103. doi: 10.1016/j.pbiomolbio.2020.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys. 1995;28:171–193. doi: 10.1017/S003358350000305X. [DOI] [PubMed] [Google Scholar]
- 10.Scott MC, Chen C-C, Mecklenburg M, Zhu C, Xu R, Ercius P, Dahmen U, Regan BC, Miao J. Electron tomography at 2.4-å;ngström resolution. Nature. 2012;483:444–447. doi: 10.1038/nature10934. [DOI] [PubMed] [Google Scholar]
- 11.Miao J, Förster F, Levi O. Equally sloped tomography with oversampling reconstruction. Phys Rev B. 2005;72:052103. doi: 10.1103/PhysRevB.72.052103. [DOI] [Google Scholar]
- 12.Brilot AF, Chen JZ, Cheng A, Pan J, Harrison SC, Potter CS, Carragher B, Henderson R, Grigorieff N. Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naydenova K, Jia P, Russo CJ. Cryo-EM with sub–1 Å specimen movement. Science. 2020;370:223–226. doi: 10.1126/science.abb7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bharat TAM, Russo CJ, Löwe J, Passmore LA, Scheres SHW. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure. 2015;23:1743–1753. doi: 10.1016/j.str.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russo CJ, Passmore LA. Progress towards an optimal specimen support for electron cryomicroscopy. Curr Opin Struct Biol. 2016;37:81–89. doi: 10.1016/j.sbi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naydenova K, Peet MJ, Russo CJ. Multifunctional graphene supports for electron cryomicroscopy. Proc Natl Acad Sci USA. 2019;116:11718–11724. doi: 10.1073/pnas.190476611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bharat TAM, Riches JD, Kolesnikova L, Welsch S, Krähling V, Davey N, Parsy ML, Becker S, Briggs JAG. Cryo-Electron Tomography of Marburg Virus Particles and Their Morphogenesis within Infected Cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farley MM, Hu B, Margolin W, Liu J. Minicells, Back in Fashion. J Bacteriol. 2016;198:1186–1195. doi: 10.1128/JB.00901-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci USA. 2012;109:E2561–E2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowall AW, Chang JJ, Freeman R, Lepault J, Walter CA, Dubochet J. Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples. J Microsc. 1983;131:1–9. doi: 10.1111/j.1365-2818.1983.tb04225.x. [DOI] [PubMed] [Google Scholar]
- 21.Melia CE, Bharat TAM. Locating macromolecules and determining structures inside bacterial cells using electron cryotomography. Biochim Biophys Acta, Proteins Proteomics. 2018;1866:973–981. doi: 10.1016/j.bbapap.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods. 2007;4:215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- 23.Zachs T, Schertel A, Medeiros J, Weiss GL, Hugener J, Matos J, Pilhofer M. Fully automated, sequential focused ion beam milling for cryo-electron tomography. Elife. 2020;9:e52286. doi: 10.7554/eLife.52286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucas BA, Grigorieff N. Quantification of gallium cryo-FIB milling damage in biological lamellae. Proc Natl Acad Sci USA. 2023;120:e2301852120. doi: 10.1073/pnas.2301852120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tacke S, Erdmann P, Wang Z, Klumpe S, Grange M, Plitzko J, Raunser S. A streamlined workflow for automated cryo focused ion beam milling. J Struct Biol. 2021;213:107743. doi: 10.1016/j.jsb.2021.107743. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Grange M, Wagner T, Kho AL, Gautel M, Raunser S. The molecular basis for sarcomere organization in vertebrate skeletal muscle. Cell. 2021;184:2135–2150.:e13. doi: 10.1016/j.cell.2021.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Grange M, Pospich S, Wagner T, Kho AL, Gautel M, Raunser S. Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin. Science. 2022;375:eabn1934. doi: 10.1126/science.abn1934. [DOI] [PubMed] [Google Scholar]
- 28.Schaffer M, Mahamid J, Engel BD, Laugks T, Baumeister W, Plitzko JM. Optimized cryo-focused ion beam sample preparation aimed atin situstructural studies of membrane proteins. J Struct Biol. 2017;197:73–82. doi: 10.1016/j.jsb.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Berger C, Premaraj N, Ravelli RBG, Knoops K, López-Iglesias C, Peters PJ. Cryo-electron tomography on focused ion beam lamellae transforms structural cell biology. Nat Methods. 2023;20:499–511. doi: 10.1038/s41592-023-01783-5. [DOI] [PubMed] [Google Scholar]
- 30.Berger C, Dumoux M, Glen T, Yee NBY, Mitchels JM, Patáková Z, Darrow MC, Naismith JH, Grange M. Plasma FIB milling for the determination of structuresin situ. Nat Commun. 2023;14:629. doi: 10.1038/s41467-023-36372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner FR, Watanabe R, Schampers R, Singh D, Persoon H, Schaffer M, Fruhstorfer P, Plitzko J, Villa E. Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat Protoc. 2020;15:2041–2070. doi: 10.1038/s41596-020-0320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melia CE, Bolla JR, Katharios-Lanwermeyer S, Mihaylov DB, Hoffmann PC, Huo J, Wozny MR, Elfari LM, Böhning J, Morgan AN, et al. Architecture of cell-cell junctions in situ reveals a mechanism for bacterial biofilm inhibition. Proc Natl Acad Sci USA. 2021;118:2109940118. doi: 10.1073/pnas.210994011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 34.Villa E, Schaffer M, Plitzko JM, Baumeister W. Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography. Curr Opin Struct Biol. 2013;23:771–777. doi: 10.1016/j.sbi.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Naydenova K, Kamegawa A, Peet MJ, Henderson R, Fujiyoshi Y, Russo CJ. On the reduction in the effects of radiation damage to two-dimensional crystals of organic and biological molecules at liquid-helium temperature. Ultramicroscopy. 2022;237:113512. doi: 10.1016/j.ultramic.2022.113512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickerson JL, Russo CJ. Phase contrast imaging with in-elastically scattered electrons from any layer of a thick specimen. Ultramicroscopy. 2022;237:113511. doi: 10.1016/j.ultramic.2022.113511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickerson JL, Lu P-H, Hristov D, Dunin-Borkowski RE, Russo CJ. Imaging biological macromolecules in thick specimens: The role of inelastic scattering in cryoEM. Ultramicroscopy. 2022;237:113510. doi: 10.1016/j.ultramic.2022.113510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMullan G, Faruqi AR, Clare D, Henderson R. Comparison of optimal performance at 300 keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy. 2014;147:156–163. doi: 10.1016/j.ultramic.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruskin AI, Yu Z, Grigorieff N. Quantitative characterization of electron detectors for transmission electron microscopy. J Struct Biol. 2013;184:385–393. doi: 10.1016/j.jsb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chreifi G, Chen S, Metskas LA, Kaplan M, Jensen GJ. Rapid tilt-series acquisition for electron cryotomography. J Struct Biol. 2019;205:163–169. doi: 10.1016/j.jsb.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenstein F, Danev R, Pilhofer M. Improved applicability and robustness of fast cryo-electron tomography data T acquisition. J Struct Biol. 2019;208:107–114. doi: 10.1016/j.jsb.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisenstein F, Yanagisawa H, Kashihara H, Kikkawa M, Tsukita S, Danev R. Parallel cryo electron tomography on in situ lamellae. Nat Methods. 2023;20:131–138. doi: 10.1038/s41592-022-01690-1. [DOI] [PubMed] [Google Scholar]
- 43.McMullan G, Faruqi AR, Henderson R. Direct Electron Detectors. Methods Enzymol. 2016;579:1–17. doi: 10.1016/bs.mie.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Zheng SQ, Egami K, Agard DA, Cheng Y. Influence of electron dose rate on electron counting images recorded with the K2 camera. J Struct Biol. 2013;184:251–260. doi: 10.1016/j.jsb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagen WJH, Wan W, Briggs JAG. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution sub-tomogram averaging. J Struct Biol. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turoňová B, Hagen WJH, Obr M, Mosalaganti S, Beugelink JW, Zimmerli CE, Kraösslich HG, Beck M. Benchmarking tomographic acquisition schemes for high-resolution structural biology. Nat Commun. 2020;11:876. doi: 10.1038/s41467-020-14535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C-C, Zhu C, White ER, Chiu CY, Scott MC, Regan BC, Marks LD, Huang Y, Miao J. Three-dimensional imaging ofdislocations in ananoparticle at atomic resolution. Nature. 2013;496:74–77. doi: 10.1038/nature12009. [DOI] [PubMed] [Google Scholar]
- 48.Lee E, Fahimian BP, Iancu CV, Suloway C, Murphy GE, Wright ER, Castaño-DÍez D, Jensen GJ, Miao J. Radiation dose reduction and image enhancement in biological imaging through equally-sloped tomography. J Struct Biol. 2008;164:221–227. doi: 10.1016/j.jsb.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Chen C-C, Scott MC, Ophus C, Xu R, Pryor A, Wu L, Sun F, Theis W, Zhou J, et al. Deciphering chemical order/disorder and material properties at the single-atom level. Nature. 2017;542:75–79. doi: 10.1038/nature21042. [DOI] [PubMed] [Google Scholar]
- 50.Pryor A, Yang Y, Rana A, Gallagher-Jones M, Zhou J, Lo YH, Melinte G, Chiu W, Rodriguez JA, Miao J. GENFIRE: A generalized Fourier iterative reconstruction algorithm for high-resolution 3D imaging. Sci Rep. 2017;7:10409. doi: 10.1038/s41598-017-09847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng Z, She Z, Zhou Q, Dong Z, Zhan F, Zhang H, Xu J, Gao Z, Dong Y. NUDIM: Anon-uniform fast Fourier transform based dual-space constraint iterative reconstruction method in biological electron tomography. J Struct Biol. 2021;213:107770. doi: 10.1016/j.jsb.2021.107770. [DOI] [PubMed] [Google Scholar]
- 52.Mastronarde DN. Dual-Axis Tomography: An Approach with Alignment Methods That Preserve Resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 53.Arslan I, Tong JR, Midgley PA. Reducing the missing wedge: High-resolution dual axis tomography of inorganic materials. Ultramicroscopy. 2006;106:994–1000. doi: 10.1016/j.ultramic.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 54.Guesdon A, Blestel S, Kervrann C, Chrétien D. Single versus dual-axis cryo-electron tomography of microtubules assembled in vitro Limits and perspectives. J Struct Biol. 2013;181:169–178. doi: 10.1016/j.jsb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Tegunov D, Xue L, Dienemann C, Cramer P, Mahamid J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Nat Methods. 2021;18:186–193. doi: 10.1038/s41592-020-01054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Candés EJ, Romberg J, Tao T. Robust uncertainty principles: exact signal reconstruction from highly incomplete frequency information. IEEE Trans Inf Theor. 2006;52:489–509. [Google Scholar]
- 57.Donoho DL. Compressed Sensing. IEEE Trans Inf Theor. 2006;52:1289–1306. [Google Scholar]
- 58.Böhning J, Bharat TAM, Collins SM. Compressed sensing for electron cryotomography and high-resolution subtomogram averaging of biological specimens. Structure. 2021;30:408–417. doi: 10.1016/j.str.2021.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leary R, Saghi Z, Midgley PA, Holland DJ. Compressed sensing electron tomography. Ultramicroscopy. 2013;131:70–91. doi: 10.1016/j.ultramic.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 60.Donati L, Nilchian M, Trépout S, Messaoudi C, Marco S, Unser M. Compressed sensing for STEM tomography. Ultramicroscopy. 2017;179:47–56. doi: 10.1016/j.ultramic.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Guay MD, Czaja W, Aronova MA, Leapman RD. Compressed Sensing Electron Tomography for Determining Biological Structure. Sci Rep. 2016;6:27614. doi: 10.1038/srep27614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saghi Z, Holland DJ, Leary R, Falqui A, Bertoni G, Sederman AJ, Gladden LF, Midgley PA. Three-Dimensional Morphology of Iron Oxide Nanoparticles with Reactive Concave Surfaces. A Compressed Sensing-Electron Tomography (CS-ET) Approach. Nano Lett. 2011;11:4666–4673. doi: 10.1021/nl202253a. [DOI] [PubMed] [Google Scholar]
- 63.Saghi Z, Divitini G, Winter B, Leary R, Spiecker E, Ducati C, Midgley PA. Compressed sensing electron tomography of needle-shaped biological specimens–Potential for improved reconstruction fidelity with reduced dose. Ultramicroscopy. 2016;160:230–238. doi: 10.1016/j.ultramic.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 64.Böhm J, Frangakis AS, Hegerl R, Nickell S, Typke D, Baumeister W. Toward detecting and identifying macromolecules in a cellular context: Template matching appliedto electron tomograms. Proc Natl Acad Sci USA. 2000;97:14245–14250. doi: 10.1073/pnas.230282097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frangakis AS, Böhm J, Förster F, Nickell S, Nicastro D, Typke D, Hegerl R, Baumeister W. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc Natl Acad Sci USA. 2002;99:14153–14158. doi: 10.1073/pnas.172520299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frangakis AS. It’s noisy out there! A review of denoising techniques in cryo-electron tomography. J Struct Biol. 2021;213:107804. doi: 10.1016/j.jsb.2021.107804. [DOI] [PubMed] [Google Scholar]
- 67.Bepler T, Kelley K, Noble AJ, Berger B. Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nat Commun. 2020;11:5208. doi: 10.1038/s41467-020-18952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu M, Singla J, Tocheva EI, Chang Y-W, Stevens RC, Jensen GJ, Alber F. De Novo Structural Pattern Mining in Cellular Electron Cryotomograms. Structure. 2019;27:679–691.:e14. doi: 10.1016/j.str.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez-Sanchez A, Kochovski Z, Laugks U, Meyer zum Alten Borgloh J, Chakraborty S, Pfeffer S, Baumeister W, LuciC V. Template-free detection and classification of membrane-bound complexes in cryo-electron tomograms. Nat Methods. 2020;17:209–216. doi: 10.1038/s41592-019-0675-5. [DOI] [PubMed] [Google Scholar]
- 70.Moebel E, Martinez-Sanchez A, Lamm L, Righetto RD, Wietrzynski W, Albert S, Lariviére D, Fourmentin E, Pfeffer S, Ortiz J, et al. Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Nat Methods. 2021;18:1386–1394. doi: 10.1038/s41592-021-01275-4. [DOI] [PubMed] [Google Scholar]
- 71.Chen M, Dai W, Sun SY, Jonasch D, He CY, Schmid MF, Chiu W, Ludtke SJ. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat Methods. 2017;14:983–985. doi: 10.1038/nmeth.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Teresa-Trueba I, Goetz SK, Mattausch A, Stojanovska F, Zimmerli CE, Toro-Nahuelpan M, Cheng DWC, Tollervey F, Pape C, Beck M, et al. Convolutional networks for supervised mining of molecular patterns within cellular context. Nat Methods. 2023;20:284–294. doi: 10.1038/s41592-022-01746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng X, Kahng A, Xue L, Mahamid J, Chang Y-W, Xu M. High-throughput cryo-ET structural pattern mining by unsupervised deep iterative subtomogram clustering. Proc Natl Acad Sci USA. 2023;120:e2213149120. doi: 10.1073/pnas.2213149120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, ZŽídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, ZŽídek A, Bates R, Blackwell S, Yim J, et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv. 2022 doi: 10.1101/2021.10.04.463034. [DOI] [Google Scholar]
- 76.Yu D, Chojnowski G, Rosenthal M, Kosinski J. AlphaPulldown—a python package for protein–protein interaction screens using AlphaFold-Multimer. Bioinformatics. 2023;39:btac749. doi: 10.1093/bioinformatics/btac749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Förster F, Hegerl R. Structure determination in situ by averaging of tomograms. Methods Cell Biol. 2007;79:741–767. doi: 10.1016/S0091-679X(06)79029-X. [DOI] [PubMed] [Google Scholar]
- 78.Scheres SHW, Melero R, Valle M, Carazo J-M. Averaging of Electron Subtomograms and Random Conical Tilt Reconstructions through Likelihood Optimization. Structure. 2009;17:1563–1572. doi: 10.1016/j.str.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Briggs JAG. Structural biology in situ — the potential of subtomogram averaging. Curr Opin Struct Biol. 2013;23:261–267. doi: 10.1016/j.sbi.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 80.Zivanov J, Otón J, Ke Z, von Kögelgen A, Pyle E, Qu K, Morado D, Castaño-Díez D, Zanetti G, Bharat TAM, et al. A Bayesian Approach to single-particle electron cryo-tomography in RELION-4.0. Elife. 2022;11:e83724. doi: 10.7554/eLife.83724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan W, Briggs JAG. Cryo-Electron Tomography and Sub-tomogram Averaging. Methods Enzymol. 2016;579:329–367. doi: 10.1016/bs.mie.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 82.Galaz-Montoya JG, Ludtke SJ. The advent of structural biology in situ by single particle cryo-electron tomography. Biophys Rep. 2017;3:17–35. doi: 10.1007/s41048-017-0040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.von Kügelgen A, Tang H, Hardy GG, Kureisaite-Ciziene D, Brun YV, Stansfeld PJ, Robinson CV, Bharat TAM. In situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer. Cell. 2020;180:348–358.:e15. doi: 10.1016/j.cell.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pfeffer S, Woellhaf MW, Herrmann JM, Förster F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun. 2015;6:6019. doi: 10.1038/ncomms7019. [DOI] [PubMed] [Google Scholar]
- 85.Carlson LA, Briggs JAG, Glass B, Riches JD, Simon MN, Johnson MC, Möller B, Grönewald K, Kraösslich HG. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bharat TAM, Riches JD, Kolesnikova L, Welsch S, Krähling V, Davey N, Parsy ML, Becker S, Briggs JAG. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bharat TAM, Kureisaite-Ciziene D, Hardy GG, Yu EW, Devant JM, Hagen WJH, Brun YV, Briggs JAG, Löwe J. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat Microbiol. 2017;2:17059. doi: 10.1038/nmicrobiol.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.von Kügelgen A, Alva V, Bharat TAM. Complete atomic structure of a native archaeal cell surface. Cell Rep. 2021;37:110052. doi: 10.1016/j.celrep.2021.110052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hutchings J, Stancheva V, Miller EA, Zanetti G. Subtomogram averaging of COPII assemblies reveals how coat organization dictates membrane shape. Nat Commun. 2018;9:4154. doi: 10.1038/s41467-018-06577-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 91.Schorb M, Briggs JAG. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy. 2014;143:24–32. doi: 10.1016/j.ultramic.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tuijtel MW, Koster AJ, Jakobs S, Faas FGA, Sharp TH. Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci Rep. 2019;9:1369. doi: 10.1038/s41598-018-37728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moser F, Pražák V, Mordhorst V, Andrade DM, Baker LA, Hagen C, Grönewald K, Kaufmann R. Cryo-SOFI enabling low-dose super-resolution correlative light and electron cryo-microscopy. Proc Natl Acad Sci USA. 2019;116:4804–4809. doi: 10.1073/pnas.1810690116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang YW, Chen S, Tocheva EI, Treuner-Lange A, Löbach S, Søgaard-Andersen L, Jensen GJ. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat Methods. 2014;11:737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaufmann R, Schellenberger P, Seiradake E, Dobbie IM, Jones EY, Davis I, Hagen C, Grünewald K. Super-Resolution Microscopy Using Standard Fluorescent Proteins in Intact Cells under Cryo-Conditions. Nano Lett. 2014;14:4171–4175. doi: 10.1021/nl501870p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.DeRosier DJ. Where in the cell is my protein? Q Rev Biophys. 2021;54:e9–e12. doi: 10.1017/S003358352100007X. [DOI] [PubMed] [Google Scholar]
- 97.Yi H, Strauss JD, Ke Z, Alonas E, Dillard RS, Hampton CM, Lamb KM, Hammonds JE, Santangelo PJ, Spearman PW, Wright ER. Native immunogold labeling of cell surface proteins and viral glycoproteins for cryo-electron microscopy and cryo-electron tomography applications. J Histochem Cytochem. 2015;63:780–792. doi: 10.1369/0022155415593323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bouchet-Marquis C, Pagratis M, Kirmse R, Hoenger A. Metallothionein as a clonable high-density marker for cryo-electron microscopy. J Struct Biol. 2012;177:119–127. doi: 10.1016/j.jsb.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Q, Mercogliano CP, Löwe J. Ferritin-Based Label for Cellular Electron Cryotomography. Structure. 2011;19:147–154. doi: 10.1016/j.str.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 100.Leapman RD, Sun S. Cryo-electron energy loss spectroscopy: observations on vitrified hydrated specimens and radiation damage. Ultramicroscopy. 1995;59:71–79. doi: 10.1016/0304-3991(95)00019-W. [DOI] [PubMed] [Google Scholar]
- 101.Aronova MA, Leapman RD. Development of Electron Energy Loss Spectroscopy in the Biological Sciences. MRS Bull. 2012;37:53–62. doi: 10.1557/mrs.2011.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wolf SG, Houben L, Elbaum M. Cryo-scanning transmission electron tomography of vitrified cells. Nat Methods. 2014;11:423–428. doi: 10.1038/nmeth.2842. [DOI] [PubMed] [Google Scholar]
- 103.Elad N, Bellapadrona G, Houben L, Sagi I, Elbaum M. Detection of isolated protein-bound metal ions by single-particle cryo-STEM. Proc Natl Acad Sci USA. 2017;114:11139–11144. doi: 10.1073/pnas.1708609114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O’Reilly FJ, Rappsilber J. Cross-linking mass spectrometry: methods and applications in structural, molecular and systems biology. Nat Struct Mol Biol. 2018;25:1000–1008. doi: 10.1038/s41594-018-0147-0. [DOI] [PubMed] [Google Scholar]
- 105.O’Reilly FJ, Xue L, Graziadei A, Sinn L, Lenz S, Tegunov D, Blötz C, Singh N, Hagen WJH, Cramer P, et al. In-cell architecture of an actively transcribing-translating expressome. Science. 2020;369:554–557. doi: 10.1126/science.abb3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bennet JL, Nguyen GTH, Donald WA. Protein–Small Molecule Interactions in Native Mass Spectrometry. Chem Rev. 2002;122:7327–7385. doi: 10.1021/acs.chemrev.1c00293. [DOI] [PubMed] [Google Scholar]
- 107.Agasid MT, Robinson CV. Probing membrane protein–lipid interactions. Curr Opin Struct Biol. 2021;69:78–85. doi: 10.1016/j.sbi.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 108.Su CC, Lyu M, Morgan CE, Bolla JR, Robinson CV, Yu EW. A’Build and Retrievé methodology to simultaneously solve cryo-EM structures of membrane proteins. Nat Methods. 2021;18:69–75. doi: 10.1038/s41592-020-01021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Esser TK, Böhning J, Fremdling P, Agasid MT, Costin A, Fort K, Konijnenberg A, Gilbert JD, Bahm A, Makarov A, et al. Mass-selective and ice-free electron cryomicroscopy protein sample preparation via native electrospray ion-beam deposition. PNAS Nexus. 2022;1:pgac153. doi: 10.1093/pnasnexus/pgac153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ochner H, Szilagyi S, Abb S, Gault J, Robinson CV, Malavolti L, Rauschenbach S, Kern K. Low-energy electron holography imaging of conformational variability of single-antibody molecules from electrospray ion beam deposition. Proc Natl Acad Sci USA. 2021;118:e2112651118. doi: 10.1073/pnas.2112651118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stevie FA. In: Introduction to Focused Ion Beams. Giannuzzi LA, Stevie FA, editors. Springer; Boston, MA: 2005. Focused Ion Beam Secondary Ion Mass Spectrometry (FIB-SIMS) [DOI] [Google Scholar]
- 112.Fletcher JS, Vickerman JC. A new SIMS paradigm for2D and 3D molecular imaging of bio-systems. Anal Bioanal Chem. 2010;396:85–104. doi: 10.1007/s00216-009-2986-3. [DOI] [PubMed] [Google Scholar]
- 113.Pillatsch L, Östlund F, Michler J. FIBSIMS: A review of secondary ion mass spectrometry for analytical dual beam focussed ion beam instruments. Prog Cryst Growth Char Mater. 2019;65:1–19. doi: 10.1016/j.pcrysgrow.2018.10.001. [DOI] [Google Scholar]
- 114.Passarelli MK, Pirkl A, Moellers R, Grinfeld D, Kollmer F, Havelund R, Newman CF, Marshall PS, Arlinghaus H, Alexander MR, et al. The 3D orbisims—label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power. Nat Methods. 2017;14:1175–1183. doi: 10.1038/nmeth.4504. [DOI] [PubMed] [Google Scholar]
- 115.Kotowska AM, Trindade GF, Mendes PM, Williams PM, Aylott JW, Shard AG, Alexander MR, Scurr DJ. Protein identification by 3D OrbiSIMS to facilitate in situ imaging and depth profiling. Nat Commun. 2020;11:5832. doi: 10.1038/s41467-020-19445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang J, Brown J, Scurr DJ, Bullen A, MacLellan-Gibson K, Williams P, Alexander MR, Hardie KR, Gilmore IS, Rakowska PD. Cryo-OrbiSIMS for 3D Molecular Imaging of a Bacterial Biofilm in Its Native State. Anal Chem. 2020;92:9008–9015. doi: 10.1021/acs.analchem.0c01125. [DOI] [PubMed] [Google Scholar]