Abstract
In European and many African, Middle Eastern and Southern Asian populations lactase persistence (LP) is the most strongly selected monogenic trait to have evolved over the last 10,000 years1. While LP selection and prehistoric milk consumption must be linked, considerable uncertainty remains concerning their spatiotemporal configuration and specific interactions2,3. We provide detailed distributions of milk exploitation across Europe over the last 9k years using c. 7,000 pottery fat residues from >550 archaeological sites. European milk use was widespread from the Neolithic period onwards but varied spatially and temporally in intensity. Surprisingly, comparison of model likelihoods indicates that LP selection varying with levels of prehistoric milk exploitation provides no better explanation of LP allele frequency trajectories than uniform selection since the Neolithic. In the UK Biobank4,5 cohort of ~500K contemporary Europeans, LP genotype was only weakly associated with milk consumption and did not show consistent associations with improved fitness or health indicators. This suggests other hypotheses on the beneficial effects of LP should be considered for its rapid frequency increase. We propose that lactase non-persistent individuals consumed milk when it became available, but that under particular conditions and microbiological milieux this was disadvantageous, driving LP selection in prehistoric Europe. Comparison of model likelihoods indicates that population fluctuations, settlement density and wild animal exploitation – proxies for these drivers – provide better explanations of LP selection than the extent of milk exploitation. These findings offer new perspectives on prehistoric milk exploitation and LP evolution.
When our ancestors started to consume the milk of domesticated ruminant animals they set in motion a sequence of events leading to the evolution of lactase persistence (LP)6 and ultimately the modern globalised dairy industry, using c.1.5 billion cattle7 as the principal milk source. Evidence for prehistoric milk exploitation, based on combined faunal and organic residue analysis, indicates considerable spatial variability. Analysis of sheep/goat age-at-death profiles suggest dairy management was concurrent with the domestication of caprines in south west Asia in the 9th millennium BCE8,9. The earliest organic residue-based evidence of dairying locates to 7th millennium BCE northwest Anatolia, and is associated with high proportions of cattle bones in faunal assemblages2. It is clear that dairy husbandry and processing technology were brought by the first farmers into Europe via the Mediterranean coast3and through the Balkans10,11 into central Europe12–15, although milk use remains undetected in Neolithic sites of Northern Greece16. In northern Europe extensive studies of organic residues in pottery and dental calculus show unequivocally that the 4th millennium BCE Neolithic inhabitants of the British Isles and Ireland were accomplished dairy farmers (e.g. refs 17–20). In prehistoric Denmark, there is evidence of milk use alongside aquatic resource processing from 4,000 BCE21. In prehistoric Finland, the region of highest milk consumption globally today7, milk use appears with the arrival of Corded Ware pottery in the early 3rd millennium BCE, although climate fluctuations may subsequently have triggered returns to hunter-gatherer-fisher subsistence strategies22,23.
Ancient DNA data indicate that most, if not all Early Neolithic people were lactase non-persistent (LNP), and that LP only reached appreciable frequencies in the Bronze and Iron Ages24–29. Such an allele frequency trajectory indicates strong selection favouring LP, and is consistent with selection starting in the Early Neolithic29,30. The main Eurasian LP-causing allele (rs4988235-A; also known as 13,910*T; ref. 31) has a highly structured geographic distribution in modern Europe32. This may reflect different strengths of selection in different regions33,34, although other explanations remain similarly plausible, such as modern frequencies being shaped by demographic processes30,35 – including an hypothesised Bronze Age Steppe origin26 (but see ref. 29) – or allele distribution reflecting its origin location30,36. A range of hypotheses to explain positive LP selection have been proposed 6,25,37–41. Among these, the calcium assimilation hypothesis33 – whereby milk supplements low vitamin D diets in low incident UVB regions, such as occur in northern Europe, is probably the most widely cited42,43. However, this hypothesis does not explain the inferred high selection strengths in lower latitude regions, such as Africa, the Middle East, Southern Europe and Southern Asia41, where milk as a source of relatively pathogen-free fluid provides a more plausible explanation37. Cubas et al.34 recently noted a latitudinal cline in the proportion of milk fats in pots in Early Neolithic Atlantic Europe, and argued that as a marker of milk usage this cline explains the modern LP distribution in the same region. Regardless of the proximal and probably regionally-specific drivers of selection on LP, milk consumption is a prerequisite44. However, LP is not accompanied at an individual level by meaningful differences in milk consumption32,38,45,46 or the risks from a range of diseases47 in modern populations, and thus its selection may underpin potential adverse consequences of milk consumption in LNP individuals in certain historically-contingent environments38.
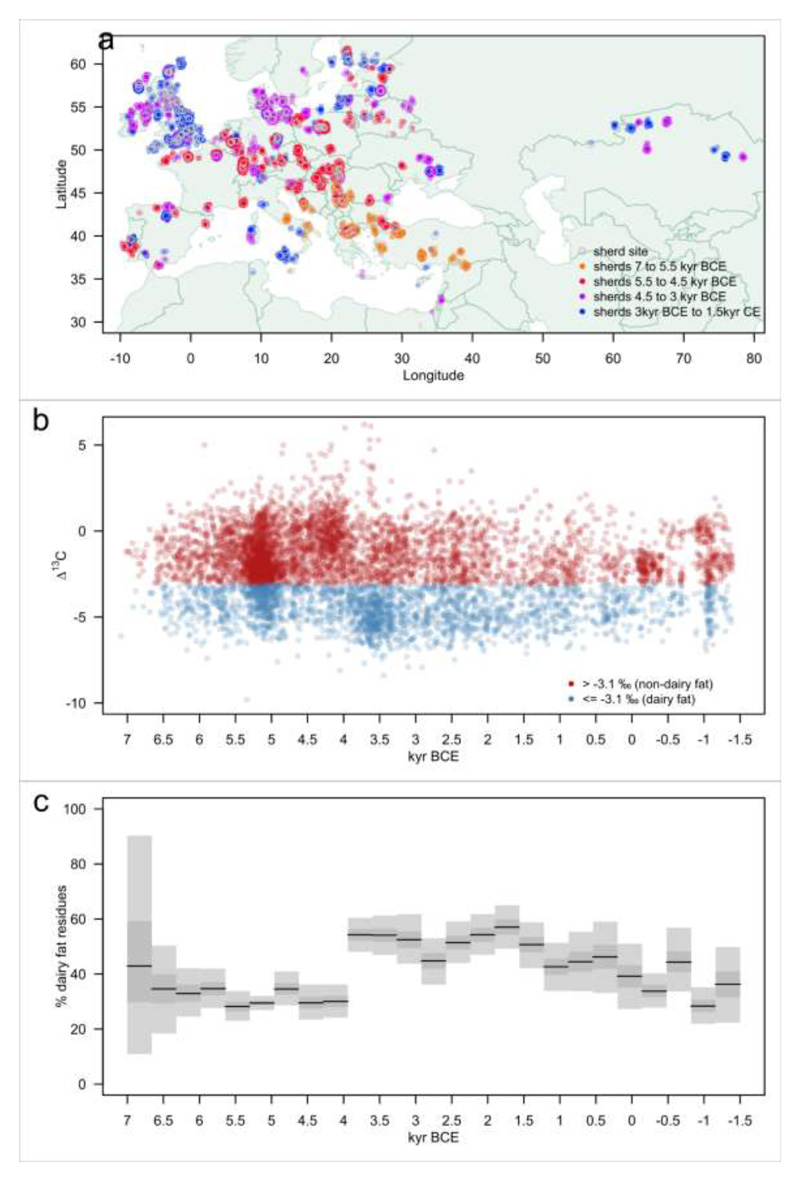
In order to build a more comprehensive picture of prehistoric milk use, and examine how well it explains selection on LP, we compiled all published instances of the occurrence of milk fat residues detected in pottery containers from 366 archaeological sites in Europe and southwest Asia. To improve spatiotemporal coverage we added newly generated organic residues data from 188 sites from various regions and periods. Overall, we utilised 6,899 animal fat residues derived from 13,181 potsherds from 826 phases from 554 sites, with associated georeferences and phase chronology including >1,000 radiocarbon dates. These data were used to generate time series depicting the frequency of milk use across prehistoric continental Europe from 7 kyr BCE to 1.5 kyr CE (Figure 1), and regional time series (Figures 2 and ED1). The high density of organic residues c.5.5 to 5 kyr BCE in Figure 1b reflects our new sampling; targeted to explore the co-evolution of dairying and LP amongst the first farmers in the central European Linearbandkeramik (LBK) culture30. If we consider the profusion of Δ13C values ≤ -3.1%o (a proxy for milk fat residues17,48,49, see Methods; Figure 1b), it becomes immediately clear that milk use was a very widespread activity across all periods in European prehistory, and at the broadest scale this was congruous with the spread of farming across the continent.
Figure 1. Geographic and temporal distribution of archaeological milk fat residues in potsherds.
(a) Coloured circles illustrate 6,899 potsherds with animal fats from 826 phases from 554 sites (grey circles). Coloured circles are jittered to prevent perfect overlay, enabling the number of sherds to be visualised. (b) Plot of Δ13C values for all 6,899 animal fat residues extracted from prehistoric potsherds through time. Each point corresponds to an individual measurement on a potsherd, with its position on the Y-axis randomly sampled from the uniform date range of the associated archaeological phase. Animal fats with Δ13C values ≤ -3.1% are defined as ruminant milk fats. Mare’s milk is not characterised using this proxy84 and would thus be defined as —non-milk”. (c) Percentage of dairy fat residues through time, calculated using all animal fat residues. Grey bars and black lines illustrate 95%, 50% Credible Intervals and Maximum a Posteriori in each time slice, using a uniform prior.
Figure 2. Regional variation in milk use in prehistoric Europe.
Interpolated time slices of the frequency of dairy fat residues in potsherds (colour hue) and confidence in the estimate (colour saturation) using 2D kernel density estimation. Bandwidth and saturation parameters were optimised using cross-validation. Circles indicate the observed frequencies at site-phase locations. The broad SE to NW cline of colour saturation at the beginning of the Neolithic illustrates a sampling bias towards earliest evidence of milk. Substantial heterogeneity in milk exploitation is evident across mainland Europe. In contrast, British Isles and Western France maintain a gradual decline across 7ka after first evidence of milk c. 5.5 ka BCE. Note that interpolation can colour some areas (particularly islands) where no data is present.
The extensive sampling and analysis of pottery has enabled us to resolve the spatiotemporal patterning of milk use in unprecedented detail (Figure 2). Importantly, this shows that milk exploitation arrived with the first farmers into the Mediterranean basin (except the region covered by modern day Greece) and continued throughout the Neolithic, whilst the later arrival of the Neolithic in Southern Britain was associated with immediate and high milk use – probably reflecting the arrival of advanced dairying populations from adjacent regions of continental Europe28 – which then gradually reduced. The prehistoric Balkans was the core region for the early intensification of milk use, which is consistent with the rise of cattle exploitation there50, yet surprisingly milk use appears largely absent from geographically adjacent Neolithic site phases located in Greece (based on analyses of >190 animal fats from >870 potsherds; however, we note that other types of containers may have been used for milk processing in this cultural context). In some regions our data suggest continuous intensive milk use, notably in western France, northern Europe and the British Isles from 5.5 kyr BCE to 1.5 kyr CE. Despite intensive sampling from central Europe (>2,800 animal fats from >6,810 potsherds), a region previously inferred to be the origin of selection on LP30, the frequency of milk use was consistently lower than for the south east, west or north of Europe throughout prehistory. The overarching picture is that, whilst dairying persisted throughout the Neolithic, its intensity fluctuated substantially in space and through time, suggesting regionally-specific instabilities in food production and cultural changes in dietary preferences. This is consistent with previous studies showing regional ‘boom and bust’ fluctuations in population density across Europe over the same period51.
LP allele (rs4988235-A) frequency trajectory estimates (Figure 3) based on published ancient DNA data from 1786 prehistoric European and Asian individuals (see subsection Data in the Ancient DNA Analyses section of the Methods), are well-described by a sigmoidal curve, as expected for a selective sweep. Curves fitted only to aDNA broadly predict modern allele frequencies32. We note that while the allele only reaches appreciable frequencies by c.2 kyr BCE29, nearly three millennia after its first detection (earliest LP individual dated to c.4,700-4,600 BCE), the dates of origin, the start of selection, the earliest observation and the reaching of appreciable frequencies of this allele are all distinct ‘events’, each possibly separated by thousands of years29. To examine if milk usage patterns can explain these allele frequency trajectories, we devised a maximum likelihood modelling approach (see Methods), whereby LP selection intensity was determined by the proportion of potsherds with dairy fat residues (as a proxy for milk use). Using simulations, we made sure that our approach had the power to detect an association if there was one, even in the presence of moderate levels of noise in our milk use patterns (see subsection Power analysis in aDNA Methods). We found that patterns of milk usage provided no better explanation (i.e. no increased probability of observed allele frequency data) than assuming uniform selection in space and through time, contra to the claims of Cubas et al.34. As an additional sensitivity test of this methodology, we tried using incident solar radiation as the main environmental variable driving LP selection intensity – in line with the calcium assimilation hypothesis33 – and obtained higher probabilities of the observed allele frequency data (Figures ED2 and ED3A, Table ED1).
Figure 3. Regional variation in prehistoric LP allele frequencies in Europe.
Observed ancient DNA data illustrated with blue and red dots showing the date and location of 3,057 LP and LNP alleles (rs4988235-A and rs4988235-G, respectively) from 1,770 ancient individuals. Grey bars and black lines show the 95%, 50% Credible Intervals and Maximum a Posteriori in time slices, using a Jeffreys prior. Slices with no observations remain blank. Blue overlay illustrates the null model of a constant selection rate through time (different in each region), using the sigmoid curve (theoretical expectation). Blue lines illustrate maximum likelihood sigmoid curves fitted directly to the observed alleles. Blue shading represents the 95%, CI, 50% CI estimated from sigmoid joint parameters using MCMC. Likelihood function for sigmoid curves are constrained by three conservative prior beliefs to avoid absurd parameter combinations for regions with sparse data: 1) constant selection is below 10% per 28 yr generation; 2) LP frequencies are <1% at 10 kyr BCE; 3) LP frequencies are >1% at 2 kyr CE. Small purple dots represent observed LP frequencies in modern populations in the same regions, as reported in Itan et al. supplementary data32, large purple dots represent a weighted average of these modern populations.
Our analysis of potsherd lipid residue and ancient LP allele data suggest that intensity of milk usage – beyond its mere presence – did not drive selection on LP. To explore other explanations for LP selection we first turned to contemporary data. The UK BioBank4,5 includes genotypic and phenotypic data from c.500,000 people aged between 37-73 years, recruited between 2006-2010 (http://biobank.ndph.ox.ac.uk/showcase). In a subset of approximately 337,000 unrelated participants who classified themselves as —white, British” and have similar genetic ancestry, LP genotypes were out of Hardy-Weinberg equilibrium (P = 1.46 x 10-18; Table ED2) which could reflect genotype-related assortative mating52 (for which there is weak evidence for LP; Table ED3), selection into a study, or population structure, amongst other influences53. We found the variant was reliably associated with a 6% increase in odds of consuming fresh cows’ milk compared with soya/no milk (95% Confidence Interval CI 1.04 - 1.09; Figure 4) and a reciprocal 6% decrease in odds of following a lactose-free diet (95% CI 0.87 – 0.99; Figure 4). Only 2.54% of dairy milk consumers were following a lactose-free diet (95% CI 2.37% - 2.71%) suggesting that consumption of lactose-depleted milk was unlikely to introduce strong bias on the genetic association with cows’ milk intake. However, around 92% (95% CI 91.5% - 92.2%) of genetically LNP participants consume fresh cows’ milk (Table 1) and among milk drinkers the LP allele was associated with a small 0.05 SD increase in daily milk servings (95% CI 0.01 – 0.08). Similar results of 0.05 SD increase in total dairy servings/day per LP allele (95% CI 0.04 – 0.07) were obtained from a large meta analysis54 of European origin individuals not included in the UK Biobank. This suggests LP has only a small effect on milk consumption, which is at odds with selection hypotheses of nutritional advantages from milk. Consistent with these results, some non-European countries with very low levels of LP have been importing milk in vast quantities in recent years as part of a more general adoption of Western diets.
Figure 4. Lactase persistence variant association with health outcomes.
Variant associations with continuous (SD unit) or binary (OR/HR) outcomes adjusted for age, sex, and top 40 genetic principal components. Unrelated estimates were produced using an unrelated white British subset of UK Biobank (n = 336,988). Within-family estimates were produced using 40,273 siblings from 19,521 families. Within-family estimates of all cause-mortality, flavoured milk intake yesterday, and milk intake yesterday could not be determined. IGF-1, insulin-like -growth factor 1. OR, odds ratio. HR, hazard ratio. Lactose-free diet (derived from field 20086 from the UK Biobank); Cows’ milk consumer (derived from field 1418); Mortality (derived from field 40007); Body Mass Index (field 21001); Father’s age at death (field 1807); Flavoured milk intake yesterday (field 100530); Heel bone mineral density (field 78); IGF-1 (field 30770); Milk intake yesterday (field 100520); Mother’s age at death (field 3526); Number of children fathered (field 2405); Number of live births (field 2734); Standing height (field 50); Vitamin D (field 30890).
Table 1. Association of lactase persistence genotype and dairy milk intake.
Unadjusted genotype association with self-reported milk preference (non-dairy, i.e. soya and never/rarely have milk, vs dairy milk; derived from field 1418) in an unrelated white British subset of UK Biobank. The variant genotypes: GG, GA and AA at locus LCT -13910 produce non-lactase persistence (GG) and lactase persistence (GA/AA) phenotypes. Association P value was produced using the chi-square test.
| Genotype | Non-milk drinkers | Milk drinkers | Total | P | ||
|---|---|---|---|---|---|---|
| GG – inferred LNP | 1,646 | 8.1% | 18,604 | 91.9% | 20,250 | |
| GA – inferred LP | 8,374 | 6.8% | 114,821 | 93.2% | 123,195 | |
| AA – inferred LP | 12,672 | 6.7% | 176,914 | 93.3% | 189,586 | |
| Total | 22,692 | 6.8% | 310,339 | 93.2% | 333,031 | 8.5 x 10-14 |
China, for example, has very low LP frequencies32 and milk was rarely consumed in the early 20th century55 but has increased more than 25-fold over the past five decades56. These findings bring into question the widely held belief that selection against LNP was the result of detrimental effects of milk consumption in otherwise healthy individuals; notably through inducing stomach cramps, diarrhoea, and flatulence41,42,57–59. One possible explanation for the small effect of LP on milk intake in contemporary data is the use of mealtime lactase supplements which reduce symptoms of lactose intolerance. However, these have only become widely available recently and UK Biobank participants would not have had access to them for most of their life.
We examined phenotypic associations of genetically-predicted LP (Figure 4) to investigate potential pathways through which selection could have occurred. The related calcium assimilation and vitamin D hypothesis33 would anticipate LP to be associated with higher vitamin D (-3.33 x 10-3 SD [95% CI - 9.22 x 10-3, 2.56 x 10-3]) concentration and bone mineral density (3.31 x 10-3 SD [95% CI -5.52 x 10-3, 0.01]) due to its positive relationship with milk consumption54,60,61. However, these estimates are very close to the null, and the large sample size puts considerable constraints on the magnitude of effects that could exist.
Milk consumption has been hypothesised to increase circulating insulin-like growth factor I62 (IGF-1), leading Wiley et al.63 to propose a model of LP selection driven by fitness advantages of IGF-1 acting to increase body size and lower age of sexual maturation. However, there were no meaningful differences in IGF-1 (-2.66 x 10-3 SD [95% CI -8.25 x 10-3, 2.93 x 10-3]). Nevertheless, LP allele presence was associated with 0.01 SD (95% CI 0.01, 0.02) higher body mass index (BMI), as previously reported54,60,61, but not with height (6.37 x 10-4 SD [95% CI -3.22 x 10-3, 4.49 x 10-3]) consistent with other studies well controlled for population stratification64,65. Whilst statistically robust and replicating what has been observed in other large samples54,60,61, the BMI effect is of small magnitude, and congruent with a small difference in calorie intake consequent of milk consumption.
LP allele association with all-cause mortality (1.00 HR [95% CI 0.97, 1.03]), mother’s age of death (-0.01 SD [95% CI -0.01, -3.03 x 10-5]) and father’s age of death (2.35 x 10-4 SD [95% CI -6.27 x 10-3, 6.74 x 10-3]), were all close to the null. These results suggest milk has little or no adverse health effect when consumed by LNP adults in a contemporary population. Another possible driver of selection is the influence of the LP allele on fertility, but we found no meaningful effect of the LP allele presence on number of live births (2.52 x 10-3 SD [95% CI -0.01, 0.01]) or number of children fathered (5.73 x 10-3 SD [95% CI -2.51 x 10-3, 0.01]).
Aside from dietary change, a number of other factors are likely to have influenced fecundity and mortality following the establishment of farming communities in Europe, including increased population and settlement density66, increased mobility67, proximity to animals, frequent crop failure, famine and population collapse51, and general poor hygiene and sanitation. Most, if not all, of these factors are likely to have increased infectious disease loads, particularly zoonoses (~61% of known and ~75% of emerging human infectious disease today come from animals68,69). Given the widespread prehistoric exploitation of milk shown here (Figure 2), and its relatively benign effects in healthy LNP individuals today, we propose two related mechanisms for the evolution of LP. Firstly, as postulated by Sverrisdóttir et al.25, the detrimental health consequences of high lactose food consumption by LNP individuals would be acutely manifested during famines, leading to high but episodic selection favouring LP. This is because lactose-induced diarrhoea can shift from an inconvenient to a fatal condition in severely malnourished individuals70, and high lactose (i.e. unfermented) milk products are more likely to be consumed when other food sources have been exhausted71. This we name the ‘crisis mechanism’, which predicts that LP selection pressures would have been greater during times of subsistence instability. A second mechanism relates to the increased pathogen loads – especially zoonoses – associated with farming and increased population density and mobility67. Mortality and morbidity due to pathogen exposure would have been amplified by the otherwise minor health effects of LNP individuals consuming milk – particularly diarrhoea – due to fluid loss and other gut disturbances, leading to enhanced selection for LP38. We name this ‘chronic mechanism’, which predicts that LP selection pressures would have increased with greater pathogen exposure. Crucially, proxies are available for some of the drivers of these hypothesised mechanisms, permitting us to explore the extent to which they explain LP selection, and so allele frequency trajectories, using our maximum likelihood modelling approach. For the ‘crisis mechanism’ we use fluctuations in population size (residual fluctuations detrended for overall background growth) as a proxy for malnutrition exposure. For the ‘chronic mechanism’, we use temporal variation in settlement density as a proxy for pathogen exposure. Both proxies are constructed using an extensive database comprising >110,000 14C dates from >27,000 sites across the European and Mediterranean area. Model likelihoods (i.e. the probabilities of the allele frequency data; Figure ED2) indicate that settlement density and population fluctuations both provide significantly (accounting for multiple testing via Benjamini-Hochberg, see Table ED1) better explanations of LP allele frequency trajectories in prehistoric western Eurasia than spatiotemporally uniform selection (Figure ED3B-C), or milk use, providing support for both the ‘crisis’ and ‘chronic’ mechanisms (i.e. the LP allele data are 284 times or 689 times more probable under models of settlement density or population fluctuations, respectively, driving selection, than a model of constant selection).
We also considered the proportion of domestic and wild animals in faunal assemblages – constructed using >1,000,000 NISP (number of identified specimens) of 17 main meat-bearing taxa from 1,093 phases from 825 sites50,72 – as explanatory variables for LP allele frequency trajectories. However, it is not clear if exploiting more domestic animals would increase pathogen exposure, as domesticates tend to live in closer proximity to humans than wild animals73, or if exploiting more wild animals would increase pathogen exposure, as greater pathogen diversity is expected for wild animals74,75. Furthermore, a shift from domestic to wild animal exploitation might result from the economic instabilities associated with the “crisis mechanism’51,76. Interestingly, the proportion of wild rather than domestic animals in archaeological assemblages provided higher model likelihoods (i.e. the LP allele data are 34 times more probable under a model of wild animal consumption driving selection, than a model of constant selection; Figure ED3D). This could be interpreted as support for either the “crisis’ or ‘chronic’ mechanisms, or be argued to counter the ‘chronic’ mechanism if prehistoric pathogen exposure was mainly from domesticates.
The prevailing narrative for the coevolution of dairying and LP has been a virtuous circle mechanism where LP frequency increased through the nutritional benefits and avoidance of negative health costs of milk consumption, facilitating an increasing reliance on milk that further drove LP selection. Our findings suggest a different picture. Milk consumption did not gradually grow throughout the European Neolithic from initially low levels, but rather was widespread at the outset in an almost entirely LNP population. We show that the scale of prehistoric milk use does not help to explain European LP allele frequency trajectories, and so selection intensities. Furthermore, we show that LP status has little impact on modern milk consumption, mortality or fecundity, and milk consumption has little or no detrimental health impact on contemporary healthy LNP individuals.
Instead, we find support for two related hypotheses: that LP selection was driven episodically and acutely by famine, and/or on a more continuous basis by synergies between pathogen exposure and the otherwise benign consequences of milk consumption in LNP individuals38. We propose that these mechanisms would have applied in the disease and malnutrition-prone environment existing during the period of rapid increase in LP frequency but would not be expected to apply outside of these circumstances. This historical contingency is supported by the substantial increase in consumption of milk in contemporary China, where the vast majority of the population are LNP77,78. In contemporary populations LP genotype associates with aspects of the gut microbiome79, and we postulate that post-weaning childhood diarrhoeal disease mortality could have been increased in LNP individuals drinking milk. Over the period in which LP prevalence increased, the ratio of late childhood (5-18 years) to early childhood (2-5 years) mortality increased80. The “crisis’ and “chronic’ mechanisms are, of course, not mutually exclusive, nor do they exclude other LP selection mechanisms, especially outside western Eurasia. We also test modern midday solar insolation incident on a horizontal surface81, and our maximum likelihood approach does indeed provide support for the calcium assimilation hypothesis33, although it should be noted that solar insolation is strongly correlated with latitude, and may therefore be confounded by factors influencing the spatial spread of the LP allele, such as its origin location, ‘allele surfing’30,35, or large-scale population movements27. Other plausible hypotheses for selection favouring LP in Europe, such as milk as a relatively pathogen-free fluid37, milk allowing earlier weaning and thus increased fertility82, milk galacto-oligosaccharide benefits for the colonic microbiome39,40, or higher efficiencies of calorie production from dairy farming83 are more difficult to assess. However, we note that the best proxy for all of these processes is milk consumption level, which itself provides no better explanation for LP allele frequency trajectories than constant selection through time. Therefore, the spatiotemporal patterns of LP selection in ancient Europeans do not reflect variation in their milk consumption. Instead, they map latitude, population volatility and settlement density; proxies for environmental conditions, subsistence economics and pathogen exposure, respectively. We note that – albeit in different configurations – these factors remain key drivers of human morbidity and mortality today.
Methods
Lipid residue analyses
Data, lipid extraction and analysis
Our lipid residue analysis draws on our SQL database of 13,181 archaeological potsherds from 554 sites, of which 6,899 sherds have yielded animal fat residues from georeferenced sites and dated phases (826 phases from 554 sites). This analysis represents c.30% new data, and c.70% collated from previously published studies1–87.
Lipid residue analyses and interpretations were based on established protocols88,89. Lipids were extracted from 1-3 g of cleaned potsherd and analysed using GC and GC-MS to determine their molecular composition. Compound-specific δ13C values of fatty acids were determined using Agilent Industries 7890A gas chromatograph coupled to an Isoprime 100 isotope ratio mass spectrometer (GC-C-IRMS) for extracts identified as animal fats. Samples were introduced via a split/splitless injector in splitless mode onto a 50 m x 0.32 mm fused silica capillary column coated with a HP-1 stationary phase (100% dimethylpolysiloxane, Agilent, 0.17 μm). The GC oven temperature programme was set to hold at 40 °C for 2 min, followed by a gradient increase to 300 °C at 10 °C min-1, the oven was then run isothermally for 10 min. Helium was used as a carrier gas and maintained at a constant flow of 2 mL min-1. The combustion reactor consisted of a quartz tube filled with copper oxide pellets which was maintained at a temperature of 850 °C. Each sample was run at least in duplicate. Instrument stability was monitored by running a fatty acid methyl ester standard mixture every 2 or 4 runs. The δ13C values are the ratios 13C/12C and expressed relative to the Vienna Pee Dee Belemnite, calibrated against a CO2 reference gas injected directly in the ion source as two pulses at the beginning of each run. Instrumental precision was 0.3 %o. Data processing was carried out using Ion Vantage software (version 1.5.6.0, IsoPrime).
Δ13C proxy for milk exploitation
The δ13C values of the C16:0 and C18:0 fatty acids from more than 170 modern ruminant adipose and dairy fats taken from animals raised in Europe, Eurasia and Africa, reveal a consistent difference in the δ13C values of the C18:0 fatty acid, which relate to the different biosynthetic origins of this fatty in the two fat types6,90. The result is that δ13C values of the C18:0 fatty acid in milk is always depleted compared to the C18:0 fatty acid in ruminant adipose fats by on average 2.3%o6. This difference allows archaeological ruminant fats extracted from pottery to be assigned as either milk or adipose fats.
Initial reference fats were taken from animals grazing in a pure C3 plant dominated pasture in the UK and archaeological investigations of dairying in historically pure C3 regions can be achieved by comparisons with the raw δ13C values, e.g. refs 6,90. However, in regions where C4 plants are present and other environmental factors, e.g. high aridity, affect the raw δ13C values, the Δ13C values (i.e. Δ13C = δ13C18:0 - δ13C16:0) are used to remove the environmental effects, e.g. refs 91,92. For modern reference fats, ruminant milk fats are distinguished from ruminant adipose fats in displaying Δ13C values < –3.1%o, ruminant adipose fats plot between –3.1 and –0.3%o, with non-ruminant fats plotting at > –0.3%o. Individual archaeological pottery lipid extracts are assigned to fat type based on their Δ13C values using the ranges defined by the reference fats.
Animal fats extracted from potsherds are categorised as containing milk lipids if their Δ13C (= δ13C18:0 δ13C16:0) values are below the threshold -3.1%o, or non-milk otherwise92. Although this threshold choice influences the absolute proportion estimates, it has almost no influence when evaluating the fluctuations in milk use through time since the relative proportions will be largely unaffected (see Methods Fig 1 below). Indeed, even if the exact number of sherds containing dairy fat residues were known, this could not be used as a proxy for milk proportion in overall diets due to biases in pottery use. Similarly, our conclusion that milk exploitation was widespread throughout the Neolithic is also based on the observation that the proportion of sherds with dairy fat residues were similar to the Bronze Age.
Methods Fig 1. Replication of Fig 1 tile C, using different thresholds for defining sherds as containing dairy fats. Correlation coefficients between all 6 comparisons vary from 0.712 to 0.982.
Chronology
Archaeological sherds are associated in the MySQL database with specific site phases. Each phase is assigned a uniform date range using published start and end dates from archaeological site reports. If unavailable, we used calibrated93 radiocarbon dates to construct phase models in OxCal94, and used the mode of each boundary posterior distribution as start and end dates. In cases where neither of these sources were available (10% of phases), phases ‘borrow’ their chronology from the geographically closest site-phase that has the same associated culture. Date ranges of site-phases are reasonably constrained (75% less than 580 years).
UK Biobank analysis
Data
UK Biobank95,96 is a large prospective cohort study of approximately 500,000 UK participants aged 37-73 years at recruitment (2006-2010). The study collected a wide-range of measures including demographics, health status, lifestyle, cognitive function, mental and physical health and molecular data such as genetic and biomarkers. UK Biobank received ethical approval from the Research Ethics Committee (REC reference for UK Biobank is 11/NW/0382). All analyses were performed under application 16729.
Dairy consumption
Multiple measures of milk consumption were evaluated through this study.
Milk servings
Participants were asked: how many glasses/carton/ 250ml of milk (excluding milkshakes) did you consume yesterday? (field 100520) and how many glasses/carton/ 250ml of yogurt drinks, flavoured milk or milkshakes did you consume yesterday? (field 100530). These measures were only asked if participants drank some milk the previous day and therefore there was no zero servings value. To reduce missingness, mean servings were calculated from five repeated measures taken in 2009-12.
Dairy milk preference
Participants were asked at baseline: What type of milk do you mainly use? (field 1418). From this variable binary non-dairy vs dairy consumption was derived: soya and never/rarely have milk vs full cream, semi-skimmed and skimmed.
Lactose-free diet
Participants were asked: Do you routinely follow a special diet? Respondents answering —Yes” to diet for lactose intolerance on any of the five repeated measures undertaken 2009-12 were compared against participants answering —No” to all non-missing responses.
Genetic data
We focused on the major European lactase persistence variant located upstream of the coding gene promotor (rs4988235, LCT -13910*T)97. The lactase genotype showed strong departure from Hardy-Weinberg equilibrium (HWE) in UK Biobank white British participants (Table ED2; x^2=77.31; P=1.46 x 10-18) using the HardyWeinberg R-package (1.6.3) (ref. 98). This finding could arise from non-random mating, clinical ascertainment bias, genotyping error, or population stratification/admixture99. The latter was mitigated as far as reasonably possible using a homogenous population which was clearly effective when considering the more extreme HWE violation in “any other white background’ group (Table ED2; x^2=422.52; P=6.9 x 10-94). HWE violation was also observed in gnomAD100 which contains genotype frequencies on 32,263 non-Finnish Europeans using exome/whole genome sequencing (Table ED2; x^2=235.75; P=3.32 x 10-53). Sequencing and array technologies have distinct error profiles and therefore technical artefact is an unlikely explanation.
Lactase gene variant crude association with dairy consumption
Unadjusted genotype frequencies stratified by dairy consumption were calculated from an unrelated white British subsample of UK Biobank prepared according to Mitchell et al.101.
Lactase gene variant association with health outcomes
Variant association with health outcomes was performed using an unrelated white British subset of UK Biobank for the main analysis and within-family estimates for sensitivity analyses. Time-to-death was estimated using a Cox proportional hazards model. All-cause mortality was derived from field age-at-death (40007). The following fields within the UK Biobank were used: Body Mass Index (21001); Father’s age at death (1807); Heel bone mineral density (78); IGF-1 (30770); Mother’s age at death (3526); Number of children fathered (2405); Number of live births (2734); Standing height (50); Vitamin D (30890). All models assumed additive inheritance and were adjusted for age, sex and top 40 genetic principal components. Within-family estimates were produced using panel regression as previously described102. All outcomes were collected at baseline.
Estimate of assortative mating on dairy consumption
To determine if milk consumption influences mate choice and therefore invalidates HWE assumptions99, we estimated the spousal LP genetic correlation (Table ED3) adjusted for age, the first ten genetic principal components, partner age, and partner first ten genetic principal components. Under an additive linear model there was very weak evidence in support of assortative mating on lactase persistence (0.004 mean increase in partner LP genotype per increase in individual LP genotype [95% CI -0.002, 0.011]). However, a single allelic copy of LP is sufficient to produce the observable phenotype (milk intake; Table 1 main text) and therefore we considered a dominant inheritance model (imputed variant dosage capped at one; rs4988235 GG vs GA & AA). Using logistic regression, we found milk consumers were at 10% increased odds of selecting a milk drinking partner (mean increase of 1.10 OR [95% CI 1.00, 1.22] partner having one or more copies of LP among LP individuals). The effect was consistent under sensitivity conditions additionally adjusting for top 40 genetic principal components (mean increase of 1.11 OR [95% CI 1.00, 1.22] partner having one or more copies of LP among LP individuals) reducing the possibility partner selection was driven entirely by population stratification.
We also found a weak effect of the variant on the number of children fathered (0.01 SD [95% CI 0.00, 0.01]; Figure 4, main text) consistent with the variant improving overall fitness which could in-part explain the positive selective pressure on the locus and observed violation of HWE.
Spouse-pairs were derived and validated in a European subsample of UK Biobank (n = 47,459) supplied by Dr Laurence Howe, MRC IEU103. Analyses were restricted to spousal-pairs with complete data (n = 46,560).
All regression models were implemented in R (v3.6.1).
Ancient DNA Analyses
Data
We collated published BAM files from 2,999 ancient Eurasian individuals104–154, out of which 1,786 had the European LP single nucleotide polymorphism (SNP) covered at least once (read depth: span 1 to 128-fold, average 7-fold). All genomes came aligned to the GRCh37/hg19 human reference assembly, but for the Tyrolean Iceman (hg18). Relevant geographic and temporal annotation was extracted from David Reich’s aDNA compendium v44.3 (ref. 155), see Figure 3 for a representation of the full dataset. For the joint analyses of genetic and ecological data, we restricted this dataset to those individuals spatiotemporally overlapping the ecological data (four geographic regions: ‘British Isles’, ‘Rhine-Danube axis’, ‘Mediterranean Europe’, and the ‘Baltic region’; 8000-2500 BP), leaving 2,162, out of which 1,293 had the lactase locus covered at least once (read depth: span 1 to 128-fold, average 7-fold).
We extracted the pileup at the position of the European LP SNP (rs4988235; in hg19: chromosome 2, position 136608646; hg18: chromosome 2, position 136325116) with default quality cutoffs using Samtools156. For sites with read depth at least two, we called the diploid genotype with the highest likelihood, for sites covered only once, only a single allele was called. Exceptions are individuals PEN001_real2 (ref. 144) and Klei10 (ref. 122), which each have one out of four reads with the derived allele. However, as discussed in the respective publications, the ages of the samples suggest the derived alleles are rather the consequence of post-mortem damage, and we therefore consider both of them as homozygous ancestral.
Likelihood of an ecological proxy variable driving LP fitness advantage
Many phenomena have the potential to influence LP allele frequency trajectories in each of our geographic regions, including selection, migration, genetic drift and population structure. We explore just one of these factors in isolation – selection – in order to test if the observed archaeological allele frequencies are best explained purely under a null model of constant selection, or might be partly explained by a model whereby the intensity of selection varies through time in accordance with a hypothesised ecological driver. While selection strength cannot be observed directly, population genetic models describe its effect on expected allele frequencies. Given the set of ecological proxy variables that we hypothesise may act as selective agents we can ask if fluctuating selection models based on these variables fit the genetic data better than a null model of constant selection strength. If a fluctuating model fits best, we may infer that the ecological proxy is linked to the true selective agent.
We obtain a likelihood equation in the following way. Say m(t) is a time series over time t, with m(t) ∈ [0,1]. We parameterise its effect on selection strength s as , where b ∈ [0, 1] is the maximal selection coefficient modulated by a factor that depends on the time series and an exponent a ∈ [0, 1]. Note that with a = 1 the model simplifies to one of constant selection strength.
As selection on lactase is strong, we assume a deterministic allele frequency trajectory, ignoring random drift. We assume a generation time of 28 years. In case of an additive model for selection—corresponding to relative fitnesses 1 + 2s, 1 + s, and 1 for homozygous derived, heterozygous, and homozygous ancestral genotypes respectively—we approximate the expected allele frequency after t generations (divide by generation time to scale to years) by with an additional model parameter y0 for the initial allele frequency at time point □0 (here set at 8000 BP). This avoids the challenges of estimating the date of the LP mutation by assuming it was present in the population at □0 at some unknown frequency, y0. Moreover, this avoids modeling the lowest frequency part of the allele trajectory where drift is strongest.
In most cases, m(t) will be piecewise constant over time windows in the order of a hundred years, meaning that s(t) is piecewise constant as well, and allele frequencies can be computed in between these boundaries with the equation above setting the initial frequency of later time intervals to the last one of the preceding time interval.
The dominant model—corresponding to relative fitnesses 1 + s, 1 + s, and 1 for homozygous derived, heterozygous, and homozygous ancestral genotypes respectively—is considered here as well – as the lactase persistence phenotype has been shown to be dominant. It requires numerical optimisation of an approximation as no exact expression for y(t) as a function of y0, s and t exists. y(t) can be obtained by keeping y0, s and t constant and adjusting y(t) until equation is satisfied157. As above, this allows us to compute expected deterministic allele frequency trajectories for piecewise constant selection strengths. Note that due to the parametrisation of fitness, the dominant selection coefficient has to be multiplied by a factor 2 to be comparable to the additive model.
For an expected allele frequency trajectory over the entire time range under one of the above inheritance models, the log likelihood given k observed ancestral alleles at time points ti and n derived alleles observed at time points tj is computed as . We were able to construct ecological proxy variables for four distinct regions, with different time series m(t) and therefore different expected allele frequency trajectories. In order to analyse these regions jointly, log-likelihoods were summed over all four regions, to result in a single optimal parameter combination across all regions.
For a given time series m(t) we optimised the model parameters y0 (initial allele frequency at time point 0), a and a (with a = 1 fixed for the constant selection strength model) using the “JDEoptim’ function from the “DEoptimR’ library158 in R159. We chose the best parameter combination from six independent runs, three with and three without initialising JDEoptim with the parameters found for the constant selection model.
Finally, statistical model comparison was performed by computing □-values obtained from a likelihood ratio test accounting for the different number of parameters between constant and fluctuating models (two and three respectively). We controlled the false discovery rate in the multiple testing of different models with the Benjamini-Hochberg procedure, i.e. ordering the □-values, □1 ≤ □2 ≤, … ≤ □□ and declare a test of rank □ non-significant at level δ if .
Power analysis
We analysed the power of our approach to estimate parameters and detect ecological proxies linked to selection strength via simulations. We chose to focus on milk exploitation, in particular to assess the robustness of our finding that milk is unlikely to drive selection on the LP allele.
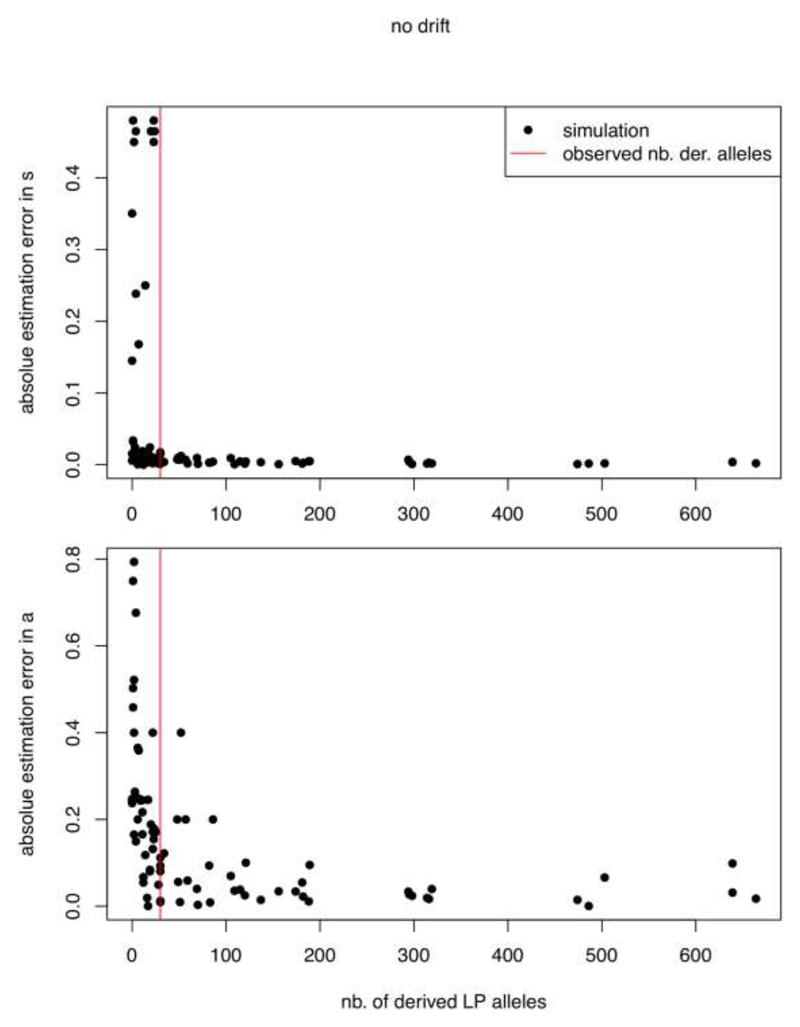
First, we simulated genotypes under nine different model parameter combinations, with three replicates per combination, sampling a matching number of alleles (one or two) at the corresponding time points in the real data. Figure ED4 shows the results for all combinations of □ ∈ {0.02,0.035,0.05} and □ ∈ {0.2,0.6,0.8}, with an initial frequency y0 fixed at 0.005. We observe that both parameters can be estimated accurately when □ > 0.02 or □ > 0.25. Models with small values for □ or □ are most challenging as both lead to small selection coefficients on average (small values □ for lead to very few bursts of high selection over a background of otherwise weak selection, see for example Figure ED3B) and therefore a generally low number of sampled derived alleles. Figure ED5 illustrates this phenomenon, where absolute errors in the estimates of □ and □ for three replicate simulations per parameter combination above, repeated independently for three different initial frequencies □0 ∈ {0.0005,0.005,0.01}, are plotted against the number of simulated derived alleles. High estimation errors are exclusively found when few derived alleles are simulated, as the latter are responsible for important likelihood differences between models with appreciable LP allele frequencies. The number of derived alleles in our dataset seems to be high enough to estimate □ with high and □ at least with intermediate accuracy. Besides the estimation of actual model parameters, we quantified the power to select a model with milk as a driver over a constant selection model. For the same set of simulations underlying Figure ED5, we show the likelihood differences between a model of constant selection and one where milk exploitation drives selection strength on the derived LP allele in Figure ED6. With less than 30 derived alleles, which is the number we observe in the actual data, only 20% of the likelihood differences pass the significance threshold, compared to 81% for simulations with at least the same number of derived alleles (51% overall), where we restricted the set of simulations only to those with an estimated parameter □ above 0.95 (values close to one effectively lead to constant selection models and therefore a likelihood difference close to zero). We are therefore confident that if milk acted as a driver of selection strength in our data we would at least find a meaningfully increased likelihood.
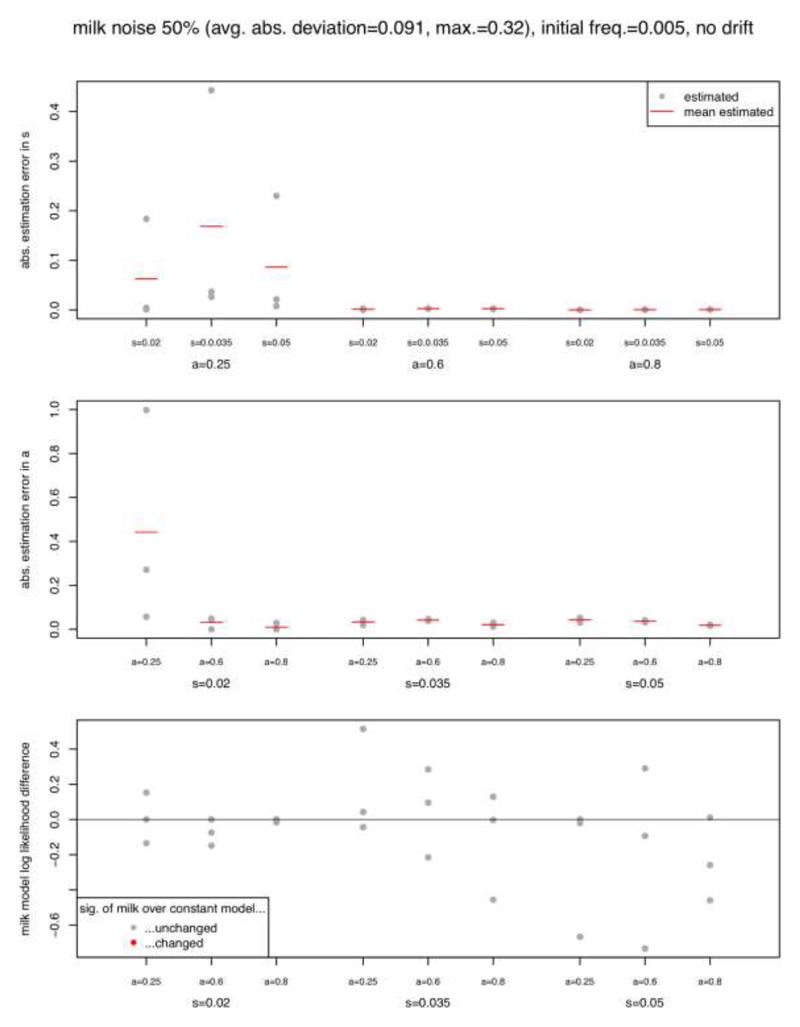
Second, we assessed the impact of noise in the ecological proxies, here exemplarily and most importantly in the context of this study on milk exploitation. To simulate inaccuracies, we multiplied each constant piece of the milk exploitation proxy by a random number uniformly drawn from the interval [0.5, 1.5], leading to a ‘noisy’ version which differs from the original values on average by 0.091 (corresponding to ~11% of the milk variable’s full range from 0 to ~0.8; maximum deviation 0.32). Figure ED7 compares parameter estimates and likelihoods with the values found in the corresponding simulation shown in Figure ED4. While there are outliers with noticeable deviation when □ = 0.25 (most challenging to distinguish from a constant selection model as discussed above), the remaining parameter estimates are barely affected, and none of the likelihoods change across the significance threshold. Therefore, we consider our approach robust against moderate levels of noise in the milk exploitation proxy, and more generally in other time series.
Lastly, we sought to quantify the effect that drift—which we are not explicitly modelling—may have on our inferences. We repeated the experiments shown in Figures ED4-ED6, however, simulating allele frequency trajectories with fluctuating selection and drift as the basis from which alleles are drawn with the sampling strategy described above that emulates our real aDNA data (drift simulations only with initial frequency □0 = 0.005). We used the ‘learnPopGen’ R library160, and simulated two levels of drift by setting the effective population size to □□ ∈ {1,000,10,000} (generation time implemented by extending allele frequencies per generation to piecewise constant frequencies lasting 28 years). We observe that the ability to accurately infer □ is only mildly affected, whereas the effect on is stronger (Figure ED8). This is in line for example with Jewett et al.161, where the authors find limited benefit to explicitly model drift for the inference of selection coefficients. Plotting the same data differently shows that the relationship with the number of simulated derived alleles continues to hold, although the accuracy □ of estimates drop with stronger drift even at high numbers of simulated derived alleles (Figure ED9). Importantly, the ability to detect significant likelihood differences in the presence of drift is not affected (Figure ED10), as detailed in Table ED4. Hence, we consider our modelling robust against mild levels of drift and a deterministic model appropriate, especially as selection coefficients are known to have been high on the derived LP allele and as our geographic regions are large enough to warrant high effective population sizes.
In conclusion, our approach performs well at estimating parameters, especially when the number of derived alleles is at least that in the real aDNA data we curated for this paper, and most importantly has the power to select a fluctuating over a constant selection model, even in the presence of moderate drift and noise in ecological driver variables.
Ecological Proxy Variables: hypothesised drivers of LP selection
We generate five ecological variables as proxies for hypothesised drivers of LP selection. Each variable is generated for four geographic polygons which overlap spatially and temporally with our ancient genetic data (‘British Isles’, ‘Baltic region’, ‘Rhine-Danube axis’ and ‘Mediterranean Europe’). We make no assumption about the direction of the possible relationship between each variable (V) and the selection coefficient - i.e. if they are positively or negatively correlated. Therefore, we also generate a set of inverse variables, which can be notionally considered as a reflection, for example a variable that ranges between 0 and 1 will have an inverse variable between 1 and 0.
Radiocarbon dates
Two of the five ecological variables (Residential density, and fluctuations in population levels) were constructed directly by utilising an extensive database comprising >110,000 14C dates which we compiled from over 1,000 published sources including CalPal162, BANADORA163, RADON164, EUROEVOL165, Wales and Borders radiocarbon database166, Scottish Radiocarbon Database167, Balsera et al.168 and the Palaeolithic Europe Database169, which provided the majority of radiocarbon dates. A summary of the radiocarbon dates used within our four polygons can be found at https://github.com/AdrianTimpson/2020-03-03523A and a full list of sources is available on request.
Residential density (clustering)
Under the assumption that dwelling proximity increases the potential for the spread of disease in a population, our ‘chronic’ mechanism hypothesises that periods and regions of greater residential density would tend to suffer more from diseases, providing greater selection on LP. We construct a residential density statistic (RDS) from the site locations of our radiocarbon database. The RDS is influenced neither by the number of archaeological sites (which is heavily biased by differential sampling, and a general increase through the Neolithic from population growth), nor by geographic structure in the site locations (caused by features such as islands and mountains). Instead, our RDS uses the distribution of pairwise distances between all sites in a region as a null expectation, and then calculates how the distribution of pairwise distances in each time slice deviates from this expected distribution. We achieve this by first generating the Empirical Cumulative Distribution Function (ECDF) of pairwise distances of all sites (ECDFall), then for a single time slice we generate the ECDF of pairwise distances of sites that fall within that time slice (ECDFt). We then calculate the RDS in that slice as sum(ECDFt – ECDFall). This is repeated for each time slice, giving a vector of RDS values. Given there is substantial chronological uncertainty for each site, we also use associated radiocarbon dates to generate a Summed Probability Distribution (SPD) after calibrating through IntCal13 (ref. 93). We use the 99% quantiles of this SPD as a conservatively wide date range for the site. We then uniformly sample a date from this range, in order to assign each site to just one time slice. The entire process is repeated 500 times to generate 500 RDS vectors. This allows CIs to be generated that incorporate the chronological uncertainty in the dataset, and we use the mean of these 500 replicates as the final time series in the LP model.
Animal exploitation (proportion of wild or domestic)
Fluctuations between domestic and wild animal exploitation for meat have the potential to inform on various economic, social and food production changes. For example, we might expect an increase in the proportion of domesticates to be associated with greater animal husbandry, and increased contact with animals could lead to a greater probability of zoonotic diseases, and therefore greater selection. Conversely, we might expect periods of famine from failure of agriculture to result in a return to exploiting wild resources, thus a decrease in domestic animals might correlate with periods (and regions) with greater selection. We extract NISP counts of the main meat bearing taxa for domestic (Bos taurus, Sus scrofa domesticus, Ovis aries, Capra hircus, Ovis aries / Capra hircus, Equus caballus) and wild (Bos primigenius, Sus scrofa scrofa, Cervus elaphus, Alces alces, Rupicapra rupicapra, Cervidae, Oryctolagus cuniculus, Lepus europaeus, Lagomorpha, Equus ferus) from the EUROEVOL170 and OSSK171 databases. Therefore, for a given time slice the ‘domesticates’ and ‘wild’ NISP counts are used to calculate a proportion of domesticates.
Fluctuations in population levels
We utilise our radiocarbon database and the R package ADMUR directly model the underlying broadscale population dynamics as the combination of exponential population growth with power law taphonomic loss. A Summed Probability Distribution (SPD) was also generated, and the residual difference between the short term fluctuations of the SPD and the long term trend of the model was used as the detrended proxy for short term fluctuations in human presence through time. This was achieved independently for each of the four geographic polygons, after all radiocarbon dates were calibrated through the intcal20 curve93 Finally, we normalise these residuals to a range between 0 and 1 globally, by scaling each variable by the smallest and largest values found across all polygons. In all cases to adjust for ascertainment bias, radiocarbon dates in a sitephase were first aggregated and normalised, so that the total probability mass for each site-phase was 1, no matter how many radiocarbon dates at a site-phase.
Solar insolation
We calculate mean annual solar insolation at 0.5 by 0.5 degree resolution using modern data (1983 to 2013) from NASA’s midday insolation incident on a horizontal surface172 (W/m2). Values at each grid point are then used to calculate a single mean value for each of the four geographic polygons. Therefore unlike the previous ecological variables that each comprise three time-series, solar insolation (and its inverse) are not time variable. No doubt insolation would have fluctuated over the study period, but the magnitude of this variation is trivial compared to the variation between polygons. These values are then normalised to a range between 0 and 1 by dividing by the Earth’s single greatest mean annual solar insolation value of 867.90 W /m2.
Time-series plots and interpolation maps
We account for three sources of uncertainty when estimating frequencies from count data (LP and LNP alleles, potsherds with and without dairy fat residues, domestic and wild species NISP). Within any time-slice we have multiple site-phases, each of which has integer counts of presence and absence. Sample sizes vary hugely between sites (potsherds between 1 and 121, NISP between 1 and 61,497), and our objective is to fairly estimate the ‘underlying’ frequencies of these count data across all these site-phases. We ensure each site contributes fairly to this estimate, by weighting by the precision of each site’s frequency estimate (a function of sample size n, where the weight is defined by ).
In addition, to fairly incorporate in our estimate the uncertainty both from small sample size and chronology, we adopt a three-stage sampling process in our estimate, which is repeated 5,000 times in order to also find the Maximum a Posteriori (MAP) and Credible Intervals. The first stage randomly samples a single frequency from a Beta distribution, where the two shape parameters are the observed presence and absence counts in the site-phase. This includes an additional one count of each as a uniform prior. The second stage randomly samples a date from the site-phase’s date range. This then allows us to assign each phase to exactly one time-slice. The third stage then calculates the weighted average frequency of the site-phases (in each time-slice), weighted according to their sample size as explained above. Where a phase spans two (or more) time slices, its contribution to each slice is weighted accordingly.
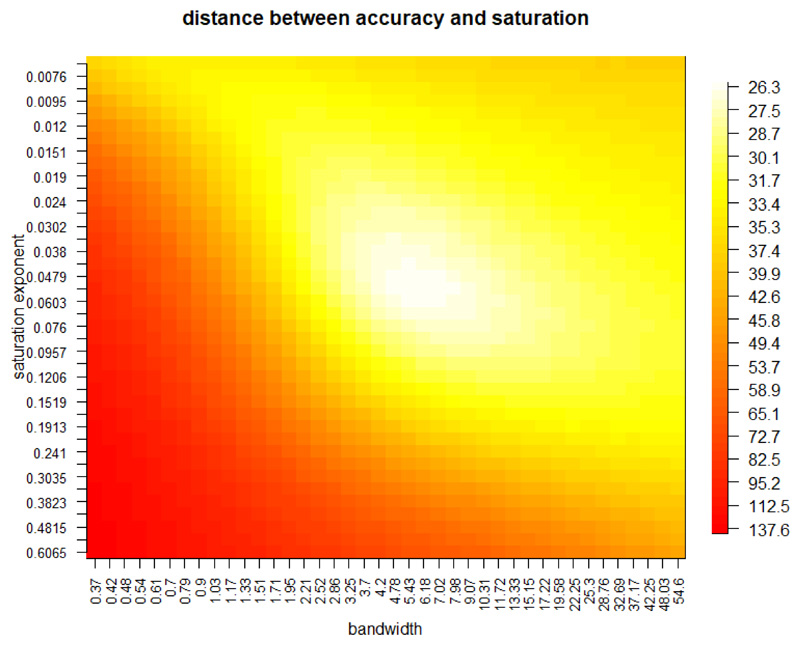
Interpolated maps of the frequency of dairy fat residues (Fig 2) were achieved with a novel kernel density approach that generates both the interpolated frequencies using colour hue, and confidence in the interpolation using colour saturation. We constructed a 2D Gaussian kernel for each site-phase, the magnitude of which was weighted by the precision of the estimate, using ) where n is the number of sherds that have yielded animal fats. The frequency of dairy residues at any geographic point was then calculated as the average of the proportion of sherds containing dairy fat residues over the total number of sherds yielding residues at a given site-phase, weighted by the probability density function (pdf) of each site-phase’s kernel at that point. Therefore, the influence each site-phase has on any interpolated point on the map depends primarily on its geographic distance from that point, but is also influenced by the error on its frequency estimate. This second influence is small in comparison, since the most extreme difference in the relative contribution of sites is only threefold (0.909 where n = 121 compared to 0.293 where n = 1). The combined surface of 2D Gaussian kernels (for all site-phases) generated during the interpolation then determines the colour saturation, to illustrate geographic regions where there is an absence of data. Conceptually this approach describes the idealised process of shooting paintballs at a grey map. A paintball corresponds to the information content at a site-phase, such that its central position is determined by the site’s latitude and longitude; hue is determined by the dairy fat frequency; and size of paintball is determined by the precision. Two further parameters are required. Firstly, the bandwidth of the kernel, which determines the spread of each paintball. Secondly, a saturation exponent which determines the hue of each paintball (the amount of information content). Both parameters were estimated using a cross-validation approach that partitioned the data into 80% training and 20% test, repeated 1,000 times. For each replicate, the training partition was used to generate interpolated maps (given a proposed bandwidth and saturation exponent), and a comparison was made at the locations of the test partition between the predicted values and the observed values. This comparison utilised a summary statistic that accounts for both the interpolated dairy residue frequencies, and the confidence (saturation). A good answer (predicted frequency close to the observed frequency) is better than an uncertain prediction (low saturation) which is better than a bad answer, therefore we seek to maximise saturation where predicted frequencies are accurate, and minimise saturation where predicted frequencies are poor. Therefore our summary statistic was the magnitude of the difference between saturation and prediction accuracy, which was minimised when bandwidth = 6.18 and the saturation exponent = 0.0538, see methods Fig. 2.
Methods Fig. 2. Bandwidth and saturation exponent parameters for interpolation plots were selected using a cross validation grid search, partitioning the observed data into an 80% training set and 20% test set. 1,000 random partitions were performed for each parameter pair, to generate a reliable summary statistic (distance between accuracy and saturation). Best estimates selected were bandwidth = 6.18 and the saturation exponent = 0.0538.
Extended Data
Figure ED1. Regional fluctuations in milk use throughout European prehistory.
Percentage of milk fats through time, calculated using all animal fat residues. Grey bars and black lines illustrate 95%, 50% CI and MAP in each time slice, using a uniform prior.
Figure ED2. Summary of model selection results for the tested ecological time series.
Inverse solar insolation, fluctuations in population level, and residential density yield models significantly better than a null model of constant selection. See Table ED1 for corresponding parameter estimates, and multiple testing correction (no change in the set of significant models). Abbreviations: assimilation (assi.), inverse (inv.), fluctuation (fluc.)
Figure ED3. A-D—Most likely models fitted to four ecological proxy variables yielding likelihoods significantly better than a constant selection model.
Although LP is generally thought of as a dominant trait, we only show the additive model results as the parameter estimates barely differ. Abbreviations: inverse (inv.), population (pop.), fluctuation (fluc.)
Figure ED4. Parameter estimation accuracy without drift.
True and inferred parameters □ and □ for three replicates of each of nine parameter combinations, simulated based on the milk exploitation ecological proxy and with initial allele frequency. □0 = 0.005. Abbreviations: frequency (freq.)
Figure ED5. Parameter estimation accuracy in relation to the number of derived alleles simulated without drift.
The simulations shown in Figure ED4 were repeated with altered initial allele frequencies □0 ∈ {0.0005,0.01}, and the entire set including □0 = 0.005. plotted as a function of the number of simulated derived alleles. The vertical line indicates the 30 derived alleles present in our aDNA dataset. Abbreviations: number (nb.), derived (der.)
Figure ED6. Likelihood differences between constant and fluctuating selection model driven by milk exploitation in relation to the number of derived alleles simulated without drift.
Same simulations as in Figure ED5. Note that parameter optimisation on simulated data was not initialised with the parameters found for the constant model, which can lead to likelihoods of the milk model to fall below the one of the former. Abbreviations: number (nb.), estimate (est.), derived (der.)
Figure ED7. Effect of noise in milk exploitation ecological proxy on parameter estimates and likelihood differences.
For each simulation presented in Figure ED4, optimisation was repeated with a noisy version of the milk exploitation variable (see Method section on aDNA analysis, subsection ‘Power analysis’ for details on how noise was simulated), and the effect on parameter estimates and likelihood differences quantified. Abbreviations: average (avg.), absolute (abs.), maximum (max.), frequency (freq.), significance (sig.)
Figure ED8. A-B—Parameter estimation accuracy with drift.
Same experiment as presented in Figure ED4, but with drift at two levels (□□ ∈ {1,000, 10,000}) affecting the simulated allele frequency trajectories. Abbreviations: average (avg.), absolute (abs.), frequency (freq.), significance (sig.)
Figure ED9. A-B—Parameter estimation accuracy in relation to the number of derived alleles simulated with drift.
Replotting of data from Figure ED8 as a function of the number of simulated derived alleles. Abbreviations: average (avg.), absolute (abs.), frequency (freq.), significance (sig.)
Figure ED10. A-B—Likelihood differences between constant and fluctuating selection model driven by milk exploitation in relation to the number of derived alleles simulated with drift.
Same simulations as in Figure ED8 and ED9. Note that parameter optimisation on simulated data was not initialised with the parameters found for the constant model, which can lead to likelihoods of the milk model to fall below the one of the former. Abbreviations: average (avg.), absolute (abs.), frequency (freq.), significance (sig.)
Table ED1. Model selection results for the tested ecological time series.
Columns 5-7 show the maximum likelihood parameter estimates. The last columns list the quantities needed for multiple testing correction by control of the false discovery rate via Benjamini-Hochberg procedure. The null hypothesis of constant selection is rejected at significance level δ, here δ = 0.02, in favor of a fluctuating selection model when the □-value of the test with rank □ is below where nine is the total number of hypothesis tests. Due to the parametrisation of fitness, the dominant selection coefficient has to be multiplied by a factor 2 to be comparable to the additive models (see Methods section on ancient DNA analysis for details). Abbreviations: number (nb.), parameters (par.), logarithmic (log.), likelihood ratio test (LRT), inverse (inv.), fluctuation (fluc.)
| Model | nb. of par. | log. likelihood | LRT p-value | ŝ | â | rank ; | reject H0? | |
|---|---|---|---|---|---|---|---|---|
| constant, additive | 2 | -138.699464119656 | NA | 0.0013 | 0.022 | NA | NA | NA |
| milk, additive | 3 | -138.699464119656 | 0.999999809769497 | 0.0013 | 0.022 | 1 | 8; 0.018 | False |
| insolation, additive | 3 | -138.699464119656 | 1 | 0.0013 | 0.022 | 1 | 9; 0.02 | False |
| insolation inv., additive | 3 | -131.702289227844 | 0.000183360816035128 | 9e–04 | 0.045 | 0.503 | 1; 0.002 | True |
| population fluc., additive | 3 | -132.204402436158 | 0.000313138336841865 | 0.001 | 0.147 | 0.259 | 2; 0.004 | True |
| population fluc. inv., additive | 3 | -138.699464062158 | 0.999729429294902 | 0.0013 | 0.022 | 1 | 7; 0.016 | False |
| cluster, additive | 3 | -133.04565281483 | 0.000771895423444998 | 7e–04 | 0.036 | 0.729 | 3; 0.007 | True |
| cluster inv., additive | 3 | -138.699464057882 | 0.999719547722535 | 0.0013 | 0.022 | 1 | 6; 0.013 | False |
| domesticates, additive | 3 | -138.699464057103 | 0.99971778486365 | 0.0013 | 0.022 | 1 | 5; 0.01 | False |
| wild, additive | 3 | -135.062356151726 | 0.00699514207820942 | 4e–04 | 0.052 | 0.696 | 4; 0.009 | True |
| constant, dominant | 2 | -138.664174169131 | NA | 0.0012 | 0.011 | NA | NA | NA |
| milk, dominant | 3 | -138.664174150102 | 0.999844347386256 | 0.0012 | 0.011 | 1 | 8; 0.018 | False |
| insolation, dominant | 3 | -138.664174104147 | 0.999712354249303 | 0.0012 | 0.011 | 1 | 6; 0.013 | False |
| insolation inv., dominant | 3 | -131.753282965012 | 0.000200992483171354 | 8e–04 | 0.024 | 0.496 | 1; 0.002 | True |
| population fluc., dominant | 3 | -132.129289121744 | 0.0003000999665612 | 0.001 | 0.089 | 0.245 | 2; 0.004 | True |
| population fluc. inv., dominant | 3 | -138.664174093425 | 0.99968952937304 | 0.0012 | 0.011 | 1 | 5; 0.01 | False |
| cluster, dominant | 3 | -133.014356965802 | 0.000775223542268811 | 7e–04 | 0.019 | 0.724 | 3; 0.007 | True |
| cluster inv., dominant | 3 | -138.664174112268 | 0.999730927447785 | 0.0012 | 0.011 | 1 | 7; 0.016 | False |
| domesticates, dominant | 3 | -138.664174150102 | 0.999844347386256 | 0.0012 | 0.011 | 1 | 9; 0.02 | False |
| wild, dominant | 3 | -135.137507299365 | 0.00791178688664611 | 4e–04 | 0.028 | 0.687 | 4; 0.009 | True |
Table ED2. Lactase genotype frequency and test for departure from Hardy-Weinberg equilibrium. Lactase persistence genotype (LCT -13910) frequencies in UK Biobank (stratified by self-reported ethnic background, field 21000) and gnomAD v3 (accessed 28/02/2020) showing strong departure from Hardy-Weinberg equilibrium in European and East Asian populations. AF, allele frequency. N, sample size. AA, homozygous lactase persistent. GA, heterozygous lactase persistent. GG, homozygous lactase non-persistent. P, chi-squared test for departure from Hardy-Weinberg equilibrium. X^2, chi-squared statistic.
| Population | AF | N | AA | GA | GG | P | x^2 |
|---|---|---|---|---|---|---|---|
| UK Biobank | |||||||
| White, British | 0.74 | 354998 | 197789 | 133046 | 24163 | 1.46 x 10-18 | 77.31 |
| White, Irish | 0.84 | 10664 | 7533 | 2864 | 267 | 0.81 | 0.06 |
| Any other white background | 0.50 | 14739 | 4310 | 6121 | 4308 | 6.90 x 10-94 | 422.52 |
| White and Black Caribbean | 0.51 | 203 | 44 | 121 | 38 | 0.01 | 7.01 |
| White and Black African | 0.46 | 98 | 18 | 54 | 26 | 0.35 | 0.87 |
| White and Asian | 0.51 | 298 | 69 | 167 | 62 | 0.04 | 4.04 |
| Any other mixed background | 0.47 | 466 | 94 | 250 | 122 | 0.11 | 2.53 |
| Indian | 0.58 | 19 | 7 | 8 | 4 | 0.79 | 0.07 |
| Pakistani | 0.50 | 7 | 1 | 5 | 1 | 0.55 | 0.36 |
| Any other Asian background | 0.09 | 171 | 1 | 29 | 141 | 0.92 | 0.01 |
| Caribbean | 0.95 | 10 | 9 | 0 | 1 | ||
| African | 0.30 | 5 | 1 | 1 | 3 | 0.67 | 0.18 |
| Any other Black background | 0.75 | 4 | 2 | 2 | 0 | - | - |
| Chinese | 0.67 | 3 | 1 | 2 | 0 | - | - |
| Other ethnic group | 0.25 | 1562 | 199 | 385 | 978 | 9.76 x 1042 | 183.19 |
| gnomAD | |||||||
| Amish | 0.76 | 448 | 267 | 149 | 32 | 0.10 | 2.68 |
| European (non-Finnish) | 0.64 | 32263 | 13797 | 13617 | 4849 | 3.32 x 10-53 | 235.75 |
| European (Finnish) | 0.58 | 5202 | 1729 | 2543 | 930 | 0.94 | 0.01 |
| Other | 0.31 | 1074 | 128 | 408 | 538 | 3.59 x 10-04 | 12.74 |
| Latino | 0.23 | 6820 | 393 | 2370 | 4057 | 0.06 | 3.50 |
| South Asian | 0.17 | 1521 | 55 | 393 | 1073 | 0.02 | 5.82 |
| Ashkenazi Jewish | 0.13 | 1661 | 23 | 383 | 1255 | 0.35 | 0.86 |
| African | 0.12 | 20992 | 340 | 4561 | 16091 | 0.43 | 0.62 |
| East Asian | 9.58 x 10-04 | 1566 | 0 | 3 | 1563 | 1.57 x 10-39 | 173.08 |
Table ED3.
Lactase persistence genotype correlation between spousal pairs in UK Biobank. Association of individual lactase persistence genotype on spouse genotype adjusted for age, partner age, top ten genetic principal components and partner top ten genetic principal components. Sensitivity analyses were additionally adjusted for top 40 genetic principal components. Additive and dominant inheritance models were evaluated. CI, confidence interval. N, sample size. OR, odds ratio.
| Test | N | Effect | 95% CI | P | |
|---|---|---|---|---|---|
| Additive | 46,560 | 0.004 | -0.002 | 0.011 | 0.22 |
| Additive (sensitivity) | 46,560 | 0.004 | -0.003 | 0.010 | 0.26 |
| Dominant | 46,560 | OR 1.10 | 1.001 | 1.220 | 0.05 |
| Dominant (sensitivity) | 46,560 | OR 1.11 | 1.001 | 1.221 | 0.05 |
Table ED4. Proportion of significant likelihood differences between constant and fluctuating selection model driven by milk exploitation.
Proportions are given separately for models with parameter close to one, as these are generally indistinguishable from constant selection models, and contrast the proportions for simulations with less and at least the same number of derived alleles as observed in the real aDNA data. Abbreviations: frequency (freq.), estimate (est.), number (nb.), derived (der.), alleles (al.), observed (obs.)
| est. of □ < 0.95 | est. of □ ≥ 0.95 | ||||||
|---|---|---|---|---|---|---|---|
| Ne | initial freq. | all | nb. der. al. ≥ obs. | nb. der. al. < obs. | all | nb. der. al. ≥ obs. | nb. der. al. < obs. |
| NA | 0.005 | 0.63 | 0.86 | 0.3 | 0.0 | 0.0 | 0.0 |
| 10,000 | 0.005 | 0.67 | 0.86 | 0.29 | 0.25 | 0.17 | 0.0 |
| 1,000 | 0.005 | 0.64 | 0.93 | 0.125 | 0.4 | 0.5 | 0.0 |
Acknowledgements
This study was funded by the European Research Council Advanced Grant —NeoMilk” FP7-IDEAS- ERC/324202. MR-S thanks the Royal Society for funding her Dorothy Hodgkin Fellowship (DHF\R1\180064 and RGF\EA\181067). NERC (Reference: CC010) and NEIF (www.isotopesuk.org) are thanked for funding and maintenance of the GC-MS and GC-C-IRMS instruments used for this work. GDS and MSL work in the MRC Integrative Epidemiology Unit at the University of Bristol (MC_UU_00011/1). Dr Penny Bickle (University of York, UK) and Mr David Altoft are acknowledged for the sampling of some potsherds from this study. We thank Saule Kalieva and Victor Logvin (Kostanay State University, Kazakhstan), Christian Lohr (Leibniz Research Institute for Archaeology, Mainz, Germany), Jens Lüning (Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany), Ivan Pavlů (Institute of Archaeology of the Academy of Sciences of the Czech Republic), and Ralf W. Schmitz (LVR-LandesMuseum, Bonn, Germany) for providing some of the sherds presented in this study. We are grateful to Dr Karen Dwyer, teaching fellow in English grammar and research methodology at UCL, for clarifying lactase non-persistence as the correct usage over non-lactase persistence, on the basis that non qualifies persistence, even if lactase persistence is considered a compound noun. We are also grateful to Dr Laurence Howe, Senior Research Associate at the MRC IEU for providing derived spousal pairs in UK Biobank. The authors acknowledge the use of the UCL Computer Science ECON High-Performance Computing (HPC) Cluster (ECON@UCL), and associated support services, in the completion of this work. This study was supported by the NIHR Biomedical Research Centre at University Hospitals Bristol and Weston NHS Foundation Trust and the University of Bristol. The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Footnotes
Author Contribution
RPE, MGT and GDS conceived the overall study. MR-S and RPE generated novel lipid residue data. MR-S., AT, YD and MSL, acquired data, assembled new databases and undertook statistical modelling. GDS and MSL performed the UK BioBank analyses. YD, AT and MGT conceptualised the selection model likelihood analysis. YD and AT performed the selection model testing. AT devised the kernel interpolation and generated Figs 1, 2 and 3. MGT, RPE, GDS, MR-S, YD, AT and MSL wrote the paper. All other authors contributed either critical archaeological information, pottery from excavations, data of various types and expert knowledge. All authors read and approved the manuscript.
Competing interest declaration
The authors have no competing interests.
Data availability statement
Data for running the ancient DNA analyses are available from: https://github.com/ydiekmann/Evershed Nature 2022 KML files, a summary of archaeological milk residue data, ecological proxy variables, and a summary of radiocarbon dates are available from: https://github.com/AdrianTimpson/2020-03-03523A
UK Biobank data are available from https://www.ukbiobank.ac.uk/
Code availability statement
R and Python code for running the ancient DNA analyses are available from: https://github.com/ydiekmann/Evershed_Nature_2022
Open-source R Code for running the UK Biobank analyses under MIT license are available from: https://github.com/MRCIEU/lp-coevolution
R Code for the generation of Figs 1,2,3 and methods Figs 1 and 2; polygon are available from: https://github.com/AdrianTimpson/2020-03-03523A
References
- 1.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 2.Evershed RP, et al. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature. 2008;455:528–531. doi: 10.1038/nature07180. [DOI] [PubMed] [Google Scholar]
- 3.Debono Spiteri C, et al. Regional asynchronicity in dairy production and processing in early farming communities of the northern Mediterranean. Proc Natl Acad Sci USA. 2016;113:13594–13599. doi: 10.1073/pnas.1607810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins R. What makes UK Biobank special? Lancet. 2012;379:1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 5.Allen NE, Sudlow C, Peakman T, Collins R. UK Biobank Data: Come and Get It. Sci Transl Med. 2014;6:224ed224. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 6.Gerbault P, et al. Evolution of lactase persistence: an example of human niche construction. Philos Trans R Soc Lond, B, Biol Sci. 2011;366:863–877. doi: 10.1098/rstb.2010.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Food and Agriculture Organization of the United Nations (FAO) FAOSTAT. Available at http://www.fao.org/faostat/en/#data/QA. [PubMed]
- 8.Vigne J-D, Helmer D. Was milk a —secondary product” in the Old World Neolithisation process? Its role in the domestication of cattle, sheep and goats. Anthropozoologica. 2007;42:9–40. [Google Scholar]
- 9.Roffet-Salque M, Gillis R, Evershed RP, Vigne J-D. In: Hybrid communities: biosocial approaches to domestication and other trans-species relationships. Stépanoff Charles, Vigne Jean-Denis., editors. Routledge; 2018. pp. 127–143. [Google Scholar]
- 10.Gillis R, et al. Sophisticated cattle dairy husbandry at Bordusani-Popină (Romania, fifth millennium BC): the evidence from complementary analysis of mortality profiles and stable isotopes. World Archaeol. 2013;45:447–472. [Google Scholar]
- 11.Ethier J, et al. Earliest expansion of animal husbandry beyond the Mediterranean zone in the sixth millennium BC. Sci Rep. 2017;7:7146. doi: 10.1038/s41598-017-07427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salque M, et al. Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature. 2013;493:522–525. doi: 10.1038/nature11698. [DOI] [PubMed] [Google Scholar]
- 13.Gillis RE, et al. The evolution of dual meat and milk cattle husbandry in Linearbandkeramik societies. Proc Royal Soc B. 2017;284 doi: 10.1098/rspb.2017.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balasse M, Tresset A. Early weaning of Neolithic domestic cattle (Bercy, France) revealed by intra-tooth variation in nitrogen isotope ratios. J Archaeol Sci. 2002;29:853–859. [Google Scholar]
- 15.Casanova E, et al. Dating the emergence of dairying by the first farmers of Central Europe using 14C analysis of fatty acids preserved in pottery vessels. Proc Natl Acad Sci USA. doi: 10.1073/pnas.2109325118. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whelton HL, Roffet-Salque M, Kotsakis K, Urem-Kotsou D, Evershed RP. Strong bias towards carcass product processing at Neolithic settlements in northern Greece revealed through absorbed lipid residues of archaeological pottery. Quat Int. 2018;496:127–139. [Google Scholar]
- 17.Copley MS, et al. Direct chemical evidence for widespread dairying in prehistoric Britain. Proc Natl Acad Sci USA. 2003;100:1524–1529. doi: 10.1073/pnas.0335955100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cramp LJE, et al. Immediate replacement of fishing with dairying by the earliest farmers of the northeast Atlantic archipelagos. Proc Royal Soc B. 2014;281:20132372. doi: 10.1098/rspb.2013.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth J, Evershed RP. Milking the megafauna: Using organic residue analysis to understand early farming practice. Environ Archaeol. 2016;21:214–229. [Google Scholar]
- 20.Charlton S, et al. New insights into Neolithic milk consumption through proteomic analysis of dental calculus. Archaeol Anthropol Sci. 2019;11:6183–6196. [Google Scholar]
- 21.Craig OE, et al. Ancient lipids reveal continuity in culinary practices across the transition to agriculture in Northern Europe. Proc Natl Acad Sci USA. 2011;108:17910–17915. doi: 10.1073/pnas.1107202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cramp LJE, et al. Neolithic dairy farming at the extreme of agriculture in Northern Europe. Proc Royal Soc B. 2014;281:20140819. doi: 10.1098/rspb.2014.0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pääkkonen M, Holmqvist E, Bläuer A, Evershed RP, Asplund H. Diverse economic patterns in the North Baltic Sea region in the Late Neolithic and Early Metal periods. Eur J Archaeol. 2019;23:4–21. [Google Scholar]
- 24.Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci USA. 2007;104:3736–3741. doi: 10.1073/pnas.0607187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sverrisdóttir OÓ, et al. Direct estimates of natural selection in Iberia indicate calcium absorption was not the only driver of lactase persistence in Europe. Mol Biol Evol. 2014;31:975–983. doi: 10.1093/molbev/msu049. [DOI] [PubMed] [Google Scholar]
- 26.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 27.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brace S, et al. Ancient genomes indicate population replacement in Early Neolithic Britain. Nat Ecol Evol. 2019;3:765–771. doi: 10.1038/s41559-019-0871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burger J, et al. Low Prevalence of Lactase Persistence in Bronze Age Europe Indicates Ongoing Strong Selection over the Last 3,000 Years. Curr Biol. 2020;30:4307–4315.:e4313. doi: 10.1016/j.cub.2020.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Itan Y, Powell A, Beaumont MA, Burger J, Thomas MG. The origins of lactase persistence in Europe. PLoS Comput Biol. 2009;5:e1000491. doi: 10.1371/journal.pcbi.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enattah NS, et al. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 32.Itan Y, Jones B, Ingram C, Swallow D, Thomas M. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol. 2010;10:36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flatz G, Rotthauwe H. Lactose nutrition and natural selection. Lancet. 1973;302:76–77. doi: 10.1016/s0140-6736(73)93267-4. [DOI] [PubMed] [Google Scholar]
- 34.Cubas M, et al. Latitudinal gradient in dairy production with the introduction of farming in Atlantic Europe. Nat Commun. 2020;11:1–9. doi: 10.1038/s41467-020-15907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klopfstein S, Currat M, Excoffier L. The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol. 2006;23:482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- 36.Gerbault P, Moret C, Currat M, Sanchez-Mazas A. Impact of selection and demography on the diffusion of lactase persistence. PloS One. 2009;4:e6369. doi: 10.1371/journal.pone.0006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook GC, al-Torki MT. High intestinal lactase concentrations in adult Arabs in Saudi Arabia. Br Med J. 1975;3:135–136. doi: 10.1136/bmj.3.5976.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davey Smith G, et al. Lactase persistence-related genetic variant: population substructure and health outcomes. Eur J Hum Genet. 2009;17:357–367. doi: 10.1038/ejhg.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cederlund A, et al. Lactose in human breast milk an inducer of innate immunity with implications for a role in intestinal homeostasis. PloS One. 2013;8 doi: 10.1371/journal.pone.0053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibson PR. History of the low FODMAP diet. J Gastroenterol Hepatol. 2017;32:5–7. doi: 10.1111/jgh.13685. [DOI] [PubMed] [Google Scholar]
- 41.Walker C, Thomas MG. In: Lactose - Evolutionary role, healh effects, and applications. Paques Marcel, Lindner Cordula., editors. Elsevier; 2019. pp. 1–48. 1. [Google Scholar]
- 42.Simoons F. Primary adult lactose intolerance and the milking habit: a problem in biologic and cultural interrelations - II. A culture historical hypothesis. Dig Dis Sci. 1970;15:695–710. doi: 10.1007/BF02235991. [DOI] [PubMed] [Google Scholar]
- 43.McCracken RD. Lactase deficiency: an example of dietary evolution. Curr Anthropol. 1971;12:479–517. [Google Scholar]
- 44.Holden C, Mace R. Phylogenetic analysis of the evolution of lactose digestion in adults. Hum Biol. 1997;69:605–628. [PubMed] [Google Scholar]
- 45.Ingram CJE, et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum Genet. 2007;120:779–788. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- 46.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joslin SEK, et al. Association of the Lactase Persistence Haplotype Block With Disease Risk in Populations of European Descent. Front Genet. 2020;11:1346. doi: 10.3389/fgene.2020.558762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudd SN, Evershed RP. Direct demonstration of milk as an element of archaeological economies. Science. 1998;282:1478–1481. doi: 10.1126/science.282.5393.1478. [DOI] [PubMed] [Google Scholar]
- 49.Dunne J, et al. First dairying in green Saharan Africa in the fifth millennium BC. Nature. 2012;486:390–394. doi: 10.1038/nature11186. [DOI] [PubMed] [Google Scholar]
- 50.Manning K, et al. The origins and spread of stock-keeping: the role of cultural and environmental influences on early Neolithic animal exploitation in Europe. Antiquity. 2013;87:1046–1059. [Google Scholar]
- 51.Shennan S, et al. Regional population collapse followed initial agriculture booms in mid-Holocene Europe. Nat Commun. 2013;4:2486. doi: 10.1038/ncomms3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howe LJ, et al. Genetic evidence for assortative mating on alcohol consumption in the UK Biobank. Nat Commun. 2019;10:5039. doi: 10.1038/s41467-019-12424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez S, Gaunt TR, Day INM. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendelian Randomization of Dairy Consumption Working Group. Dairy Consumption and Body Mass Index Among Adults: Mendelian Randomization Analysis of 184802 Individuals from 25 Studies. Clin Chem. 2018;64:183–191. doi: 10.1373/clinchem.2017.280701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simmons JS, Whayne TF, Anderson GW, Horack HM. Global epidemiology A geography of disease and sanitation. Vol. 1 William Heineman; 1944. [Google Scholar]
- 56.Bai Z, et al. Global environmental costs of China’s thirst for milk. Glob Change Biol. 2018;24:2198–2211. doi: 10.1111/gcb.14047. [DOI] [PubMed] [Google Scholar]
- 57.Simoons F. Primary adult lactose intolerance and the milking habit: a problem in biological and cultural interrelations - I. Review of the medical research. Dig Dis Sci. 1969;14:819–836. doi: 10.1007/BF02233204. [DOI] [PubMed] [Google Scholar]
- 58.Bayless TM, Paige DM, Ferry GD. Lactose intolerance and milk drinking habits. Gastroenterology. 1971;60:605–608. [PubMed] [Google Scholar]
- 59.Szilagyi A, Walker C, Thomas MG. In: Lactose: Evolutionary role, health effects, and applications. Paques Marcel, Lindner Cordula., editors. Elsevier; 2019. pp. 113–153. [Google Scholar]
- 60.Almon R, Álvarez-Leon EE, Serra-Majem L. Association of the European lactase persistence variant (LCT-13910 C>T polymorphism) with obesity in the Canary Islands. PLoS One. 2012;7:e43978. doi: 10.1371/journal.pone.0043978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartwig FP, Horta BL, Smith GD, de Mola CL, Victora CG. Association of lactase persistence genotype with milk consumption, obesity and blood pressure: a Mendelian randomization study in the 1982 Pelotas (Brazil) Birth Cohort, with a systematic review and meta-analysis. Int J Epidemiol. 2016;45:1573–1587. doi: 10.1093/ije/dyw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin L-Q, He K, Xu J-Y. Milk consumption and circulating insulin-like growth factor-I level: a systematic literature review. Int J Food Sci Nutr. 2009;60:330–340. doi: 10.1080/09637480903150114. [DOI] [PubMed] [Google Scholar]
- 63.Wiley A. The evolution of lactase persistence: milk consumption, insulin-like growth factor I, and human life-history parameters. Q Rev Biol. 2018;93:319–345. [Google Scholar]
- 64.Campbell CD, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 65.Bergholdt HKM, Nordestgaard BG, Varbo A, Ellervik C. Milk intake is not associated with ischaemic heart disease in observational or Mendelian randomization analyses in 98 529 Danish adults. Int J Epidemiol. 2015;44:587–603. doi: 10.1093/ije/dyv109. [DOI] [PubMed] [Google Scholar]
- 66.Bocquet-Appel J-P. When the world’s population took off: the springboard of the Neolithic demographic transition. Science. 2011;333:560–561. doi: 10.1126/science.1208880. [DOI] [PubMed] [Google Scholar]
- 67.Loog L, et al. Estimating mobility using sparse data: Application to human genetic variation. Proc Natl Acad Sci USA. 2017;114:12213–12218. doi: 10.1073/pnas.1703642114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vorou R, Papavassiliou V, Tsiodras S. Emerging zoonoses and vector-borne infections affecting humans in Europe. Epidemiol Infect. 2007;135:1231–1247. doi: 10.1017/S0950268807008527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allen T, et al. Global hotspots and correlates of emerging zoonotic diseases. Nat Commun. 2017;8:1124. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rice AL, Sacco L, Hyder A, Black RE. Malnutrition as an underlying cause of childhood deaths associated with infectious diseases in developing countries. Bull World Health Organ. 2000;78:1207–1221. [PMC free article] [PubMed] [Google Scholar]
- 71.Colledge S, Conolly J, Crema E, Shennan S. Neolithic population crash in northwest Europe associated with agricultural crisis. Quat Res. 2019:1–22. [Google Scholar]
- 72.Manning K. The Cultural Evolution of Neolithic Europe. EUROEVOL Dataset 2: Zooarchaeological data. JOAD. 2016;5 [Google Scholar]
- 73.Chessa B, et al. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009;324:532. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karesh WB, Cook RA, Bennett EL, Newcomb J. Wildlife trade and global disease emergence. Emerg Infect Dis. 2005;11:1000. doi: 10.3201/eid1107.050194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chomel BB, Belotto A, Meslin F-X. Wildlife, exotic pets, and emerging zoonoses. Emerg Infect Dis. 2007;13:6. doi: 10.3201/eid1301.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schibler J, Jacomet S, Hüster-Plogmann H, Brombacher C. Economic crash in the 37th and 36th centuries cal. BC in Neolithic lake shore sites in Switzerland. Anthropozoologica. 1997;25:553–570. [Google Scholar]
- 77.He Y, Yang X, Xia J, Zhao L, Yang Y. Consumption of meat and dairy products in China: a review. Proc Nutr Soc. 2016;75:385–391. doi: 10.1017/S0029665116000641. [DOI] [PubMed] [Google Scholar]
- 78.Mak VSW. Milk craze Body, science, and hope in China. University of Hawai’i Press; 2021. [Google Scholar]
- 79.Goodrich JuliaK, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paine RR, Boldsen JL. In: The evolution of human life history School for Advanced Research Advanced Seminar Series. Hawkes K, Paine RR, editors. School of American Research Press; 2006. pp. 307–330. [Google Scholar]
- 81.Stackhouse PW, Jr, et al. POWER release 8 (with GIS applications) methodology (data parameters, sources, & validation) documentation date may 1, 2018 (all previous versions are obsolete)(data version 8.0. 1) NASA; 2018. Retrieved from https://power.larc.nasa.gov. [Google Scholar]
- 82.Lieberman M, Lieberman D. Lactase Deficiency: A Genetic Mechanism which Regulates the Time of Weaning. Am Nat. 1978;112:625–627. [Google Scholar]
- 83.Ingold T. Hunters, pastoralists and ranchers: reindeer economies and their transformations. Cambridge University Press; Cambridge: 1980. [Google Scholar]
- 84.Outram AK, et al. The earliest horse harnessing and milking. Science. 2009;323:1332–1335. doi: 10.1126/science.1168594. [DOI] [PubMed] [Google Scholar]
- 1.Breu A, Gómez-Bach A, Heron C, Rosell-Melé A, Molist M. Variation in pottery use across the Early Neolithic in the Barcelona plain. Archaeol Anthropol Sci. 2021;13:53. [Google Scholar]
- 2.Brychova V, et al. Animal exploitation and pottery use during the early LBK phases of the Neolithic site of Bylany (Czech Republic) tracked through lipid residue analysis. Quat Int. 2021;574:91–101. [Google Scholar]
- 3.Carrer F, et al. Chemical analysis of pottery demonstrates Prehistoric origin for high-altitude alpine dairying. PloS One. 2016;11:e0151442. doi: 10.1371/journal.pone.0151442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casanova E, et al. Spatial and temporal disparities in human subsistence in the Neolithic Rhineland gateway. J Archaeol Sci. 2020;122:105215 [Google Scholar]
- 5.Colonese AC, et al. The identification of poultry processing in archaeological ceramic vessels using in-situ isotope references for organic residue analysis. J Archaeol Sci. 2017;78:179–192. [Google Scholar]
- 6.Copley MS, et al. Direct chemical evidence for widespread dairying in prehistoric Britain. Proc Natl Acad Sci USA. 2003;100:1524–1529. doi: 10.1073/pnas.0335955100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copley MS, et al. Dairying in antiquity. I. Evidence from absorbed lipid residues dating to the British Iron Age. J Archaeol Sci. 2005;32:485–503. [Google Scholar]
- 8.Copley MS, Berstan R, Straker V, Payne S, Evershed RP. Dairying in antiquity. II. Evidence from absorbed lipid residues dating to the British Bronze Age. J Archaeol Sci. 2005;32:505–521. [Google Scholar]
- 9.Copley MS, et al. Dairying in antiquity. III. Evidence from absorbed lipid residues dating to the British Neolithic. J Archaeol Sci. 2005;32:523–546. [Google Scholar]
- 10.Copley MS, Evershed RP. In: Building memories: the Neolithic Cotswold long barrow at Ascott-under-Wychwood, Oxfordshire Cardiff Studies in Archaeology. Benson Don, Whittle A., editors. Oxbow Books; 2006. [Google Scholar]
- 11.Courel B, et al. Organic residue analysis shows sub-regional patterns in the use of pottery by Northern European hunter-gatherers. R Soc Open Sci. 2020;7:192016. doi: 10.1098/rsos.192016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig OE, et al. Did the first farmers of central and eastern Europe produce dairy foods? Antiquity. 2005;79:882–894. [Google Scholar]
- 13.Craig OE, Taylor G, Mulville J, Collins MJ, Parker Pearson M. The identification of prehistoric dairying activities in the Western Isles of Scotland: an integrated biomolecular approach. J Archaeol Sci. 2005;32:91–103. [Google Scholar]
- 14.Craig OE, et al. Molecular and isotopic demonstration of the processing of aquatic products in Northern European Prehistoric pottery. Archaeometry. 2007;49:135–152. [Google Scholar]
- 15.Craig O. In: Archaeology meets science - Biomolecular investigations in Bronze Age Greece. Tzedakis Yannis, Martlew Holley, Jones Martin K., editors. Oxbow Books; 2008. pp. 121–124. [Google Scholar]
- 16.Craig OE, et al. Ancient lipids reveal continuity in culinary practices across the transition to agriculture in Northern Europe. Proc Natl Acad Sci USA. 2011;108:17910–17915. doi: 10.1073/pnas.1107202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig OE, et al. Feeding Stonehenge: cuisine and consumption at the Late Neolithic site of Durrington Walls. Antiquity. 2015;89:1096–1109. [Google Scholar]
- 18.Craig-Atkins E, et al. The dietary impact of the Norman Conquest: A multiproxy archaeological investigation of Oxford, UK. PloS One. 2020;15:e0235005. doi: 10.1371/journal.pone.0235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cramp LJE, Evershed RP, Eckardt H. What was a mortarium used for? Organic residues and cultural change in Iron Age and Roman Britain. Antiquity. 2011;85:1339–1352. [Google Scholar]
- 20.Cramp LJE, et al. Neolithic dairy farming at the extreme of agriculture in Northern Europe. Proc Royal Soc B. 2014;281:20140819. doi: 10.1098/rspb.2014.0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cramp LJE, et al. Immediate replacement of fishing with dairying by the earliest farmers of the northeast Atlantic archipelagos. Proc Royal Soc B. 2014;281:20132372. doi: 10.1098/rspb.2013.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cramp LJE, et al. Regional diversity in subsistence among early farmers in Southeast Europe revealed by archaeological organic residues. Proc Royal Soc B. 2019;286:20182347. doi: 10.1098/rspb.2018.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramp LJE, Król D, Rutter M, Heyd VM, Pospieszny L. Analiza pozostalosci organicznych z ceramiki kultury rzucewskiej z Rzucewa. Pomorania Antiqua. 2019;XXVIII:245–259. [Google Scholar]
- 24.Cubas M, et al. Latitudinal gradient in dairy production with the introduction of farming in Atlantic Europe. Nat Commun. 2020;11:1–9. doi: 10.1038/s41467-020-15907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debono Spiteri C, et al. Regional asynchronicity in dairy production and processing in early farming communities of the northern Mediterranean. Proc Natl Acad Sci USA. 2016;113:13594–13599. doi: 10.1073/pnas.1607810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demirci Ö, Lucquin A, Craig OE, Raemaekers DCM. First lipid residue analysis of Early Neolithic pottery from Swifterbant (the Netherlands, ca. 4300-4000 BC) Archaeol Anthropol Sci. 2020;12:105. [Google Scholar]
- 27.Demirci Ö, Lucquin A, Cakirlar C, Craig OE, Raemaekers DCM. Lipid residue analysis on Swifterbant pottery (c. 5000-3800 cal BC) in the Lower Rhine-Meuse area (the Netherlands) and its implications for human-animal interactions in relation to the Neolithisation process. J Archaeol Sci Rep. 2021;36:102812 [Google Scholar]
- 28.Dreslerová D, et al. Seeking the meaning of a unique mountain site through a multidisciplinary approach. The Late La Téne site at Sklářské Valley, Šumava Mountains, Czech Republic. Quat Int. 2020;542:88–108. [Google Scholar]
- 29.Drieu L, et al. Chemical evidence for the persistence of wine production and trade in Early Medieval Islamic Sicily. Proc Natl Acad Sci USA. 2021;118:e2017983118. doi: 10.1073/pnas.2017983118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drieu L, et al. A Neolithic without dairy? Chemical evidence from the content of ceramics from the Pendimoun rock-shelter (Castellar, France, 5750-5150 BCE) J Archaeol Sci Rep. 2021;35:102682 [Google Scholar]
- 31.Dunne J, et al. Milk of ruminants in ceramic baby bottles from prehistoric child graves. Nature. 2019;574:246–248. doi: 10.1038/s41586-019-1572-x. [DOI] [PubMed] [Google Scholar]
- 32.Dunne J, Chapman A, Blinkhorn P, Evershed RP. Reconciling organic residue analysis, faunal, archaeobotanical and historical records: Diet and the medieval peasant at West Cotton, Raunds, Northamptonshire. J Archaeol Sci. 2019;107:58–70. [Google Scholar]
- 33.Dunne J, Chapman A, Blinkhorn P, Evershed RP. Fit for purpose? Organic residue analysis and vessel specialisation: The perfectly utilitarian medieval pottery assemblage from West Cotton, Raunds. J Archaeol Sci. 2020;120:105178 [Google Scholar]
- 34.Dunne J, et al. Finding Oxford’s medieval Jewry using organic residue analysis, faunal records and historical documents. Archaeol Anthropol Sci. 2021;13:48. [Google Scholar]
- 35.Ethier J, et al. Earliest expansion of animal husbandry beyond the Mediterranean zone in the sixth millennium BC. Sci Rep. 2017;7:7146. doi: 10.1038/s41598-017-07427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evershed RP, Copley MS, Dickson L, Hansel FA. Experimental evidence for the processing of marine animal products and other commodities containing polyunsaturated fatty acids in pottery vessels. Archaeometry. 2008;50:101–113. [Google Scholar]
- 37.Fanti L, et al. The role of pottery in Middle Neolithic societies of western Mediterranean (Sardinia, Italy, 4500-4000 cal BC) revealed through an integrated morphometric, use-wear, biomolecular and isotopic approach. J Archaeol Sci. 2018;93:110–128. [Google Scholar]
- 38.Francés-Negro M, et al. Neolithic to Bronze Age economy and animal management revealed using analyses of lipid residues of pottery vessels and faunal remains at El Portalón de Cueva Mayor (Sierra de Atapuerca, Spain) J Archaeol Sci. 2021;131:105380 [Google Scholar]
- 39.Gregg MW, Banning EB, Gibbs K, Slater GF. Subsistence practices and pottery use in Neolithic Jordan: molecular and isotopic evidence. J Archaeol Sci. 2009;36:937–946. [Google Scholar]
- 40.Gunnarssone A, Oras E, Talbot HM, Ilves K, Legzdina D. Cooking for the living and the dead: lipid analysess of Rauši settlement and cemetery pottery from the 11th-13th century. Estonian J Archaeol. 2020;24 [Google Scholar]
- 41.Heron C, et al. Cooking fish and drinking milk? Patterns in pottery use in the southeastern Baltic, 3300-2400 cal BC. J Archaeol Sci. 2015;63:33–43. [Google Scholar]
- 42.Hoekman-Sites HA, Giblin JI. Prehistoric animal use on the Great Hungarian Plain: A synthesis of isotope and residue analyses from the Neolithic and Copper Age. J Anthropol Archaeol. 2012;31:515–527. [Google Scholar]
- 43.Isaksson S, Hallgren F. Lipid residue analyses of Early Neolithic funnel-beaker pottery from Skogsmossen, eastern Central Sweden, and the earliest evidence of dairying in Sweden. J Archaeol Sci. 2012;39:3600–3609. [Google Scholar]
- 44.Krueger M, Bajčev O, Whelton HL, Evershed RP. The Neolithic in the Middle Morava Valley. 2019;3:61–76. [Google Scholar]
- 45.Manzano E, et al. An integrated multianalytical approach to the reconstruction of daily activities at the Bronze Age settlement in Peñalosa (Jaén, Spain) Microchem J. 2015;122:127–136. [Google Scholar]
- 46.Manzano E, et al. Molecular and isotopic analyses on prehistoric pottery from the Virués-Martínez cave (Granada, Spain) J Archaeol Sci Rep. 2019;27:101929 [Google Scholar]
- 47.Matlova V, et al. Defining pottery use and animal management at the Neolithic site of Bylany (Czech Republic) J Archaeol Sci Rep. 2017;14:262–274. [Google Scholar]
- 48.McClure SB, et al. Fatty acid specific δ13C values reveal earliest Mediterranean cheese production 7,200 years ago. PloS One. 2018;13:e0202807. doi: 10.1371/journal.pone.0202807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mileto S, Kaiser E, Rassamakin Y, Whelton H, Evershed RP. Differing modes of animal exploitation in North-Pontic Eneolithic and Bronze Age Societies. Sci Technol Archaeol Res. 2017;3:112–125. [Google Scholar]
- 50.Mukherjee AJ, Berstan R, Copley MS, Gibson AM, Evershed RP. Compound-specific stable carbon isotopic detection of pig product processing in British Late Neolithic pottery. Antiquity. 2007;83:743–754. [Google Scholar]
- 51.Mukherjee AJ, Gibson AM, Evershed RP. Trends in pig product processing at British Neolithic Grooved Ware sites traced through organic residues in potsherds. J Archaeol Sci. 2008;35:2059–2073. [Google Scholar]
- 52.Ogrinc N, Budja M, Potocnik D, Žibrat Gašparič A, Mlekuž D. Lipids, pots and food processing at Hocevarica, Ljubljansko barje, Slovenia. Doc Praehist. 2014;XLI:181–194. [Google Scholar]
- 53.Oras E, et al. The adoption of pottery by north-east European hunter-gatherers: Evidence from lipid residue analysis. J Archaeol Sci. 2017;78:112–119. [Google Scholar]
- 54.Oras E, et al. Social food here and hereafter: Multiproxy analysis of gender-specific food consumption in conversion period inhumation cemetery at Kukruse, NE-Estonia. J Archaeol Sci. 2018;97:90–101. [Google Scholar]
- 55.Outram AK, et al. The earliest horse harnessing and milking. Science. 2009;323:1332–1335. doi: 10.1126/science.1168594. [DOI] [PubMed] [Google Scholar]
- 56.Outram AK, et al. Horses for the dead: funerary foodways in Bronze Age Kazakhstan. Antiquity. 2010;85:116–128. [Google Scholar]
- 57.Outram AK, et al. Patterns of pastoralism in later Bronze Age Kazakhstan: new evidence from faunal and lipid residue analyses. J Archaeol Sci. 2012;39:2424–2435. [Google Scholar]
- 58.Özbal H, et al. Neolitik Bati Anadolu ve Marmara yerleşimleri çanak çömleklerinde organik kalinti analizleri. Arkeometri Sonuçglari Toplantisi. 2013;28:105–114. [Google Scholar]
- 59.Özbal H, et al. Yenikapi, Aşağipinar, Bademağaci ve Barcin Çömleklerindeorganik kalinti analizi. Arkeometri Sonuçglari Toplantisi. 2013;29:83–90. [Google Scholar]
- 60.Pääkkönen M, Bläuer A, Evershed RP, Asplund H. Reconstructing food procurement and processing in early Comb ware period through organic residues in early Comb and Jakarla ware pottery. Fennosc Archaeol. 2016;XXXIII:57–75. [Google Scholar]
- 61.Pääkkönen M, Bläuer A, Olsen B, Evershed RP, Asplund H. Contrasting patterns of prehistoric human diet and subsistence in northernmost Europe. Sci Rep. 2018;8:1148. doi: 10.1038/s41598-018-19409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pääkkönen M, Holmqvist E, Bläuer A, Evershed RP, Asplund H. Diverse economic patterns in the North Baltic Sea region in the Late Neolithic and Early Metal periods. Eur J Archaeol. 2019;23:4–21. [Google Scholar]
- 63.Papakosta V, Oras E, Isaksson S. Early pottery use across the Baltic - A comparative lipid residue study on Ertebølle and Narva ceramics from coastal hunter-gatherer sites in southern Scandinavia, northern Germany and Estonia. J Archaeol Sci Rep. 2019;24:142–151. [Google Scholar]
- 64.Pennetta A, Fico D, Lucrezia Savino M, Larocca F, Egidio De Benedetto G. Characterization of Bronze age pottery from the Grotte di Pertosa-Auletta (Italy): Results from the first analysis of organic lipid residues. J Archaeol Sci Rep. 2020;31:102308 [Google Scholar]
- 65.Piličiauskas G, et al. The Corded Ware culture in the Eastern Baltic: New evidence on chronology, diet, beaker, bone and flint tool function. J Archaeol Sci Rep. 2018;21:538–552. [Google Scholar]
- 66.Piličiauskas G, et al. Fishers of the Corded Ware culture in the Eastern Baltic. Acta Archaeol. 2020;91:95–120. [Google Scholar]
- 67.Robinson G, et al. Furness’s first farmers: evidence of Early Neolithic settlement and dairying in Cumbria. Proc Prehist. 2020:1–34. [Google Scholar]
- 68.Robson HK, et al. Diet, cuisine and consumption practices of the first farmers in the southeastern Baltic. Archaeol Anthropol Sci. 2019 doi: 10.1007/s12520-019-00804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roffet-Salque M, Evershed RP. In: Kopydlowo, stanowisko 6 Osady neolityczne z pogranicza Kujaw i Wielkopolski. Marciniak Arkadiusz, Sobkowiak-Tabaka Iwona, Bartkowiak Marta, Lisowski Mikolaj., editors. Wydawnictwo Profil-Archeo; 2015. pp. 133–142. [Google Scholar]
- 70.Roffet-Salque M, Banecki B, Evershed RP. In: Megalityczny grobowiec kultury amfor kulistych z Kierzkowa Milczący świadek kultu przodków z epoki kamienia / A Megalithic tomb of the Globular Amphora Culture from Kierzkowo in the Paluki region - a silent witness of ancestor worship from the Stone Age Biskupin Archaeological Works. Nowaczyk S, Pospieszny L, Sobkowiak-Tabaka I, editors. Archaeological Museum in Biskupin; 2017. pp. 251–266. [Google Scholar]
- 71.Roffet-Salque M, et al. In: Ludwinowo 7 - Neolithic settlement in Kuyavia Saved Archaeological Heritage 8. Pyzel Joanna., editor. Profil-Archeo Publishing House and Archaeological Studio, University of Gdansk Publishing House; 2019. pp. 301–316. 9. [Google Scholar]
- 72.Salque M, et al. New insights into the early Neolithic economy and management of animals in Southern and Central Europe revealed using lipid residue analyses of pottery vessels. Anthropozoologica. 2012;47:45–61. [Google Scholar]
- 73.Salque M, et al. Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature. 2013;493:522–525. doi: 10.1038/nature11698. [DOI] [PubMed] [Google Scholar]
- 74.Smyth J, Evershed RP. The molecules of meals: new insight into Neolithic foodways. Proc R Ir Acad. 2015;115C:1–20. [Google Scholar]
- 75.Smyth J, Evershed RP. Milking the megafauna: Using organic residue analysis to understand early farming practice. Environ Archaeol. 2016;21:214–229. [Google Scholar]
- 76.Šoberl L. Pots for the afterlife: organic residue analysis of British Early Bronze Age pottery from funerary contexts. PhD thesis, University of Bristol; 2011. [Google Scholar]
- 77.Šoberl L, Žibrat Gašparič A, Budja M, Evershed RP. Early herding practices revealed through organic residue analysis of pottery from the early Neolithic rock shelter of Mala Triglavca, Slovenia. Doc Praehist. 2008:253–260. [Google Scholar]
- 78.Šoberl L, et al. Neolithic and Eneolithic activities inferred from organic residue analysis of pottery from Mala Triglavca, Moverna vas and Ajdovska jama, Slovenia. Doc Praehist. 2014;XLI:149–179. [Google Scholar]
- 79.Spangenberg JE, Jacomet S, Schibler J. Chemical analyses of organic residues in archaeological pottery from Arbon Bleiche 3, Switzerland - evidence for dairying in the late Neolithic. J Archaeol Sci. 2006;33:1–13. [Google Scholar]
- 80.Spangenberg JE, Matuschik I, Jacomet S, Schibler J. Direct evidence for the existence of dairying farms in prehistoric Central Europe (4th mil. BC) Isotopes Environ Health Stud. 2008;44:189–200. doi: 10.1080/10256010802066349. [DOI] [PubMed] [Google Scholar]
- 81.Spataro M, et al. Production and function of Neolithic black-painted pottery from Schela Cladovei (Iron Gates, Romania) Archaeol Anthropol Sci. 2019 [Google Scholar]
- 82.Steele VJ, Stern B. Red Lustrous Wheelmade ware: Analysis of organic residues in Late Bronze Age trade and storage vessels from the eastern Mediterranean. J Archaeol Sci Rep. 2017;16:641–657. [Google Scholar]
- 83.Stojanovski D, et al. Living off the land: Terrestrial-based diet and dairying in the farming communities of the Neolithic Balkans. PloS One. 2020;15:e0237608. doi: 10.1371/journal.pone.0237608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stojanovski D, et al. Anta 1 de Val da Laje - the first direct view at diet, dairying practice and socio-economic aspects of pottery use in the final Neolithic of central Portugal. Quat Int. 2020;542:1–8. [Google Scholar]
- 85.Tarifa-Mateo N, et al. New insights from Neolithic pottery analyses reveal subsistence practices and pottery use in early farmers from Cueva de El Toro (Malagá, Spain) Archaeol Anthropol Sci. 2019 [Google Scholar]
- 86.Weber J, Brozio JP, Mჼller J, Schwark L. Grave gifts manifest the ritual status of cattle in Neolithic societies of northern Germany. J Archaeol Sci. 2020;117:105122 [Google Scholar]
- 87.Whelton HL, Roffet-Salque M, Kotsakis K, Urem-Kotsou D, Evershed RP. Strong bias towards carcass product processing at Neolithic settlements in northern Greece revealed through absorbed lipid residues of archaeological pottery. Quat Int. 2018;496:127–139. [Google Scholar]
- 88.Evershed RP, Heron C, Goad LJ. Analysis of organic residues of archaeological origin by high-temperature gas chromatography and gas chromatography-mass spectrometry. Analyst. 1990;115:1339–1342. [Google Scholar]
- 89.Correa-Ascencio M, Evershed RP. High throughput screening of organic residues in archaeological potsherds using direct methanolic acid extraction. Anal Methods. 2014;6:1330–1340. [Google Scholar]
- 90.Dudd SN, Evershed RP. Direct demonstration of milk as an element of archaeological economies. Science. 1998;282:1478–1481. doi: 10.1126/science.282.5393.1478. [DOI] [PubMed] [Google Scholar]
- 91.Evershed RP, et al. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature. 2008;455:528–531. doi: 10.1038/nature07180. [DOI] [PubMed] [Google Scholar]
- 92.Dunne J, et al. First dairying in green Saharan Africa in the fifth millennium BC. Nature. 2012;486:390–394. doi: 10.1038/nature11186. [DOI] [PubMed] [Google Scholar]
- 93.Reimer PJ, et al. IntCal13 and Marine13 radiocarbon age calibration curves 0-50,000 years cal BP. Radiocarbon. 2013;55:1869–1887. [Google Scholar]
- 94.Bronk Ramsey C. Bayesian analysis of radiocarbon dates. Radiocarbon. 2009;51:337–360. [Google Scholar]
- 95.Collins R. What makes UK Biobank special? Lancet. 2012;379:1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 96.Allen NE, Sudlow C, Peakman T, Collins R. UK Biobank Data: Come and Get It. Sci Transl Med. 2014;6:224ed224. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 97.Enattah NS, et al. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 98.Graffelman J. Exploring Diallelic Genetic Markers: The HardyWeinberg Package. J Stat Softw. 2015;64:1–23. [Google Scholar]
- 99.Rodriguez S, Gaunt TR, Day INM. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mitchell R, Hemani G, Dudding T, Paternoster L. UK Biobank Genetic Data: MRC-IEU Quality Control. University of Bristol; 2019. Version 2. [DOI] [Google Scholar]
- 102.Brumpton B, et al. Avoiding dynastic, assortative mating, and population stratification biases in Mendelian randomization through within-family analyses. Nat Commun. 2020;11:3519. doi: 10.1038/s41467-020-17117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Howe LJ, et al. Genetic evidence for assortative mating on alcohol consumption in the UK Biobank. Nat Commun. 2019;10:5039. doi: 10.1038/s41467-019-12424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Agranat-Tamir L, et al. The Genomic History of the Bronze Age Southern Levant. Cell. 2020;181:1146–1157.:e1111. doi: 10.1016/j.cell.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 106.Antonio ML, et al. Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science. 2019;366:708–714. doi: 10.1126/science.aay6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brace S, et al. Ancient genomes indicate population replacement in Early Neolithic Britain. Nat Ecol Evol. 2019;3:765–771. doi: 10.1038/s41559-019-0871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Broushaki F, et al. Early Neolithic genomes from the eastern Fertile Crescent. Science. 2016;353:499–503. doi: 10.1126/science.aaf7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brunel S, et al. Ancient genomes from present-day France unveil 7,000 years of its demographic history. Proc Natl Acad Sci USA. 2020;117:12791. doi: 10.1073/pnas.1918034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cassidy LM, et al. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc Natl Acad Sci USA. 2015;113:368–373. doi: 10.1073/pnas.1518445113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cassidy LM, et al. A dynastic elite in monumental Neolithic society. Nature. 2020;582:384–388. doi: 10.1038/s41586-020-2378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Damgaard PdB, et al. 137 ancient human genomes from across the Eurasian steppes. Nature. 2018;557:369–374. doi: 10.1038/s41586-018-0094-2. [DOI] [PubMed] [Google Scholar]
- 113.Damgaard PdB, et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science. 2018;360:eaar7711. doi: 10.1126/science.aar7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Feldman M, et al. Ancient DNA sheds light on the genetic origins of early Iron Age Philistines. Sci Adv. 2019;5:eaax0061. doi: 10.1126/sciadv.aax0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fernandes DM, et al. The spread of steppe and Iranian-related ancestry in the islands of the western Mediterranean. Nat Ecol Evol. 2020;4:334–345. doi: 10.1038/s41559-020-1102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fu Q, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.González-Fortes G, et al. Paleogenomic Evidence for Multi-generational Mixing between Neolithic Farmers and Mesolithic Hunter-Gatherers in the Lower Danube Basin. Curr Biol. 2017;27:1801–1810.:e1810. doi: 10.1016/j.cub.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.González-Fortes G, et al. A western route of prehistoric human migration from Africa into the Iberian Peninsula. Proc Royal Soc B. 2019;286:20182288. doi: 10.1098/rspb.2018.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Günther T, et al. Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haber M, et al. Continuity and Admixture in the Last Five Millennia of Levantine History from Ancient Canaanite and Present-Day Lebanese Genome Sequences. Am J Hum Genet. 2017;101:274–282. doi: 10.1016/j.ajhg.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Harney E, et al. Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nat Commun. 2018;9:3336. doi: 10.1038/s41467-018-05649-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hofmanová Z, et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc Natl Acad Sci USA. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Järve M, et al. Shifts in the Genetic Landscape of the Western Eurasian Steppe Associated with the Beginning and End of the Scythian Dominance. Curr Biol. 2019;29:2430–2441.:e2410. doi: 10.1016/j.cub.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 124.Jones ER, et al. The Neolithic Transition in the Baltic Was Not Driven by Admixture with Early European Farmers. Curr Biol. 2017;27:576–582. doi: 10.1016/j.cub.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Keller A, et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 126.Krzewińska M, et al. Ancient genomes suggest the eastern Pontic-Caspian steppe as the source of western Iron Age nomads. Sci Adv. 2018;4:eaat4457. doi: 10.1126/sciadv.aat4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lamnidis TC, et al. Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe. Nat Commun. 2018;9:5018. doi: 10.1038/s41467-018-07483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lazaridis I, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lazaridis I, et al. Genetic origins of the Minoans and Mycenaeans. Nature. 2017;548:214–218. doi: 10.1038/nature23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Linderholm A, et al. Corded Ware cultural complexity uncovered using genomic and isotopic analysis from south-eastern Poland. Sci Rep. 2020;10:6885. doi: 10.1038/s41598-020-63138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lipson M, et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 2017;551:368–372. doi: 10.1038/nature24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Malmström H, et al. The genomic ancestry of the Scandinavian Battle Axe Culture people and their relation to the broader Corded Ware horizon. Proc Royal Soc B. 2019;286:20191528. doi: 10.1098/rspb.2019.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Marcus JH, et al. Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia. Nat Commun. 2020;11:939. doi: 10.1038/s41467-020-14523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Margaryan A, et al. Population genomics of the Viking world. Nature. 2020;585:390–396. doi: 10.1038/s41586-020-2688-8. [DOI] [PubMed] [Google Scholar]
- 136.Martiniano R, et al. The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 2017;13:e1006852. doi: 10.1371/journal.pgen.1006852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mathieson I, et al. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mittnik A, et al. Kinship-based social inequality in Bronze Age Europe. Science. 2019;93:eaax6219. doi: 10.1126/science.aax6219. [DOI] [PubMed] [Google Scholar]
- 140.Narasimhan VM, et al. The formation of human populations in South and Central Asia. Science. 2019;365 doi: 10.1126/science.aat7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Olalde I, et al. A common genetic origin for early farmers from Mediterranean Cardial and Central European LBK cultures. Mol Biol Evol. 2015;32:3132–3142. doi: 10.1093/molbev/msv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Olalde I, et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018;555:190–196. doi: 10.1038/nature25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Olalde I, et al. The genomic history of the Iberian Peninsula over the past 8000 years. Science. 2019;363:1230–1234. doi: 10.1126/science.aav4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rivollat M, et al. Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers. Sci Adv. 2020;6:eaaz5344. doi: 10.1126/sciadv.aaz5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Saag L, et al. Extensive Farming in Estonia Started through a Sex-Biased Migration from the Steppe. Curr Biol. 2017;27:2185–2193.:e2186. doi: 10.1016/j.cub.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 146.Sánchez-Quinto F, et al. Megalithic tombs in western and northern Neolithic Europe were linked to a kindred society. Proc Natl Acad Sci USA. 2019;116:9469–9474. doi: 10.1073/pnas.1818037116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schuenemann VJ, et al. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat Commun. 2017;8:15694. doi: 10.1038/ncomms15694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schroeder H, et al. Unraveling ancestry, kinship, and violence in a Late Neolithic mass grave. Proc Natl Acad Sci USA. 2019;116:10705–10710. doi: 10.1073/pnas.1820210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Skoglund P, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 150.Skourtanioti E, et al. Genomic History of Neolithic to Bronze Age Anatolia, Northern Levant, and Southern Caucasus. Cell. 2020;181:1158–1175.:e1128. doi: 10.1016/j.cell.2020.04.044. [DOI] [PubMed] [Google Scholar]
- 151.Unterlander M, et al. Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nat Commun. 2017;8:14615. doi: 10.1038/ncomms14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Valdiosera C, et al. Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc Natl Acad Sci USA. 2018;115:3428–3433. doi: 10.1073/pnas.1717762115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Villalba-Mouco V, et al. Survival of Late Pleistocene Hunter-Gatherer Ancestry in the Iberian Peninsula. Curr Biol. 2019;29:1169–1177.:e1167. doi: 10.1016/j.cub.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 154.Wang C-C, et al. Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat Commun. 2019;10:590. doi: 10.1038/s41467-018-08220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reich D. Allen Ancient DNA Resource (AADR): Downloadable genotypes of present-day and ancient DNA data. 2021. V44.3. https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data.
- 156.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Felsenstein J. Theoretical evolutionary genetics. University of Washington; Seattle: 2016. [Google Scholar]
- 158.Brest J, Greiner S, Boskovic B, Mernik M, Zumer V. Self-adapting control parameters in differential evolution: A comparative study on numerical benchmark problems. IEEE Trans Evol Comput. 2006;10:646–657. [Google Scholar]
- 159.R Core Team. R: A language and environment for statistical computing. R foundation for statistical computing; 2018. Available at http://www.R-project.org/ [Google Scholar]
- 160.Revell LJ. learnPopGen: An R package for population genetic simulation and numerical analysis. Ecol Evol. 2019;9:7896–7902. doi: 10.1002/ece3.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jewett EM, Steinrücken M, Song YS. The Effects of Population Size Histories on Estimates of Selection Coefficients from Time-Series Genetic Data. Mol Biol Evol. 2016;33:3002–3027. doi: 10.1093/molbev/msw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Weninger B. CalPal-2007. Cologne radiocarbon calibration & palaeoclimate research package; 2007. http://www.calpal.de . [Google Scholar]
- 163.Galate P. BANADORA, Banque de données des dates radiocarbones de Lyon pour l’Europe et le Proche-Orient. 2011. http://www.archeometrie.mom.fr/banadora .
- 164.Hinz M, et al. RADON-Radiocarbon dates online 2012. Central European database of 14C dates for the Neolithic and the Early Bronze Age. Journal of Neolithic Archaeology. 2012 [Google Scholar]
- 165.Manning K, Colledge S, Crema ER, Shennan S, Timpson A. The Cultural Evolution of Neolithic Europe. EUROEVOL Dataset 1: Sites, Phases and Radiocarbon Data. JOAD. 2016;5 [Google Scholar]
- 166.Burrow S, Williams S. The Wales and Borders Radiocarbon Database. Amgueddfa Cymru; National Museum Wales: 2008. [Google Scholar]
- 167.Ralston I, Ashmore P. Canmore Scottish Radiocarbon Database. https://canmore.org.uk/project/919374 .
- 168.Balsera V, Diíaz-dei-Riío P, Gilman A, Uriarte A, Vicent JM. Approaching the demography of late prehistoric Iberia through summed calibrated date probability distributions (7000-2000 cal BC) Quat Int. 2015;386:208–211. [Google Scholar]
- 169.Vermeersch PM. Radiocarbon Palaeolithic Europe Database v.18. 2015. https://ees.kuleuven.be/geography/projects/14c-palaeolithic/index.html . [DOI] [PMC free article] [PubMed]
- 170.Manning K. The Cultural Evolution of Neolithic Europe. EUROEVOL Dataset 2: Zooarchaeological data. JOAD. 2016;5 [Google Scholar]
- 171.Manning K, et al. The origins and spread of stock-keeping: the role of cultural and environmental influences on early Neolithic animal exploitation in Europe. Antiquity. 2013;87:1046–1059. [Google Scholar]
- 172.Stackhouse PW, Jr, et al. POWER release 8 (with GIS applications) methodology (data parameters, sources, & validation) documentation date may 1, 2018 (all previous versions are obsolete)(data version 8.0. 1) NASA; 2018. Retrieved from https://power.larc.nasa.gov. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data for running the ancient DNA analyses are available from: https://github.com/ydiekmann/Evershed Nature 2022 KML files, a summary of archaeological milk residue data, ecological proxy variables, and a summary of radiocarbon dates are available from: https://github.com/AdrianTimpson/2020-03-03523A
UK Biobank data are available from https://www.ukbiobank.ac.uk/
R and Python code for running the ancient DNA analyses are available from: https://github.com/ydiekmann/Evershed_Nature_2022
Open-source R Code for running the UK Biobank analyses under MIT license are available from: https://github.com/MRCIEU/lp-coevolution
R Code for the generation of Figs 1,2,3 and methods Figs 1 and 2; polygon are available from: https://github.com/AdrianTimpson/2020-03-03523A