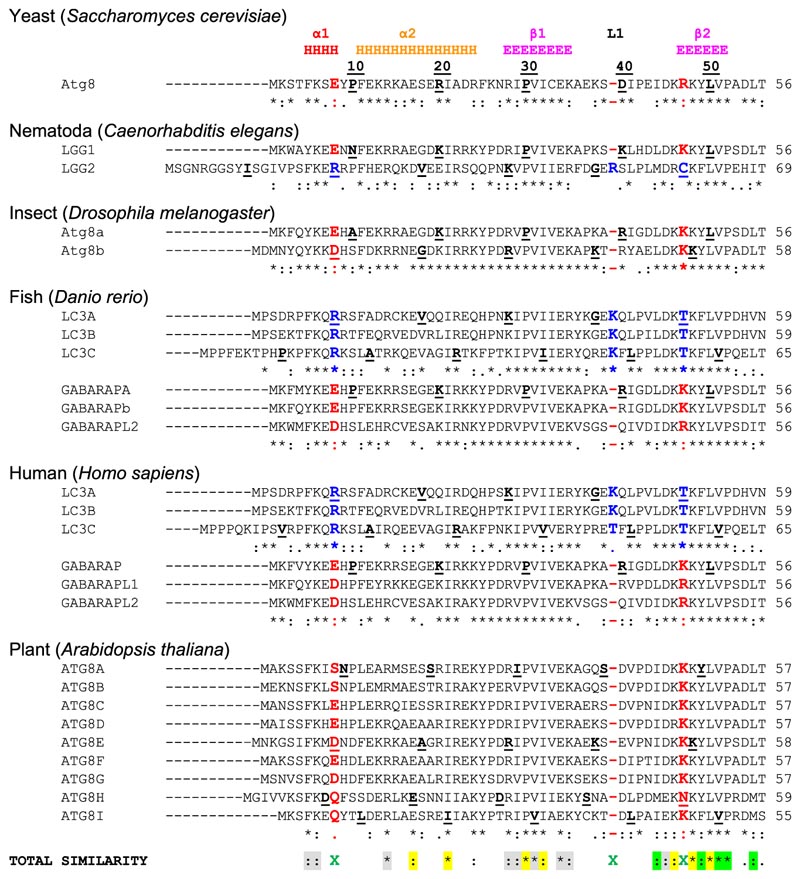
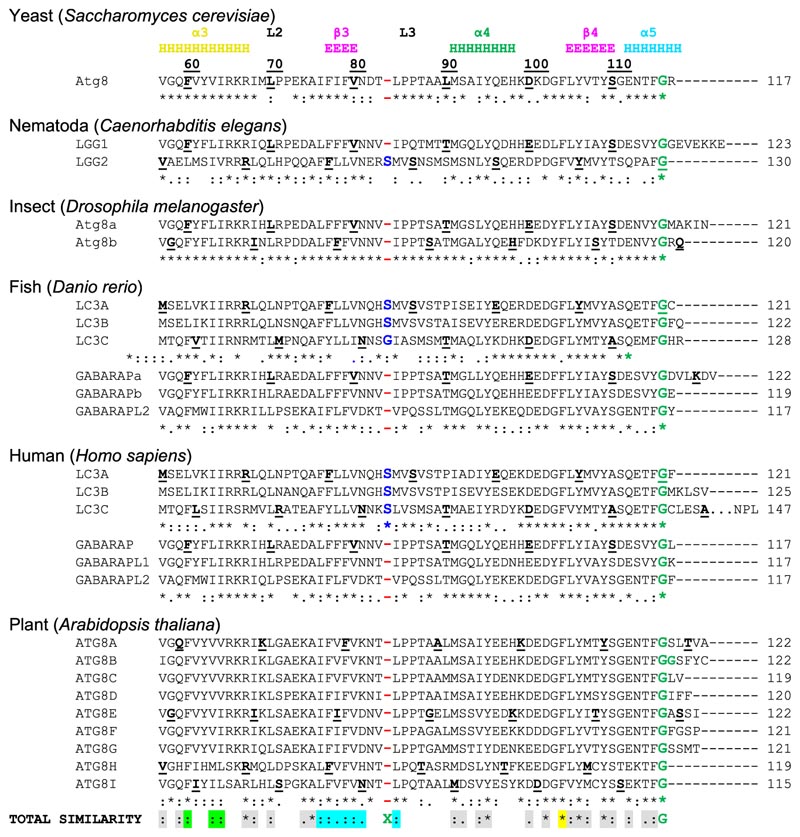
Figure 2. Sequence alignment of Atg8/LC3/GABARAP proteins.
Sequence alignment of the Atg8-family members from 6 model species – yeast (Saccharomyces cerevisiae), nematoda (Caenorhabditis elegans), insects (Drosophila melanogaster), bony fish (Danio rerio), human (Homo sapiens) and plant (Arabidopsis thaliana). Secondary structure elements from the human LC3B (PDB ID 2ZJD) are shown at the top (H – α-helices, E – β-strands, L – long loops; rainbow color-code for α-helices – red, orange, yellow, green and cyan, all β-strands are in magenta). Every tenth residue in each sequence is marked bold/underlined, the catalytic Gly is marked green. The identity scores (* for identical residues,: for very similar residues, for analogous residues, space for residues without any similarity, - for gaps) are presented below each group of the Atg8/LC3/GABARAP. For the yeast Atg8 proteins, annotated UniProt entries for 11 yeast species Atg8 sequences were aligned to generate the identity score. The residues (or their absence) separating GABARAP/Atg8 and LC3 protein subtypes are marked red and blue, respectively. The consensus string for all 38 proteins aligned is presented at the bottom of alignment (named TOTAL SIMILARITY). The residues showed conservation are grouped within the following classes: residues participating in the protein folding (grey); residues forming HP1 (yellow); residues forming HP2 (light green); and residues forming UDS (cyan). The key residues indicating LC3 and Atg8/GABARAP subtypes difference are marked by green X.